Figure 1
- ID
- ZDB-FIG-230518-10
- Publication
- Tornini et al., 2023 - linc-mipep and linc-wrb encode micropeptides that regulate chromatin accessibility in vertebrate-specific neural cells
- Other Figures
- All Figure Page
- Back to All Figure Page
|
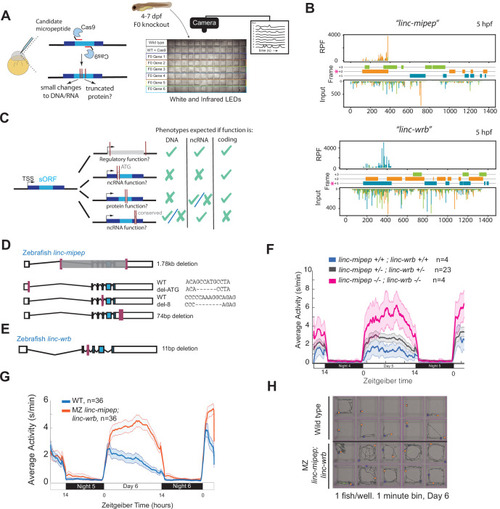
(A) Left, schematic of F0 CRISPR knockout behavioral screen. Zebrafish embryos were injected early at the one-cell embryo stage with multiple sgRNAs and Cas9 targeting the ORF of candidate micropeptides encoded within putative lincRNAs. Right, schematic of behavior screening platform. Each well of a 96-well flat bottom plate contains one zebrafish larva (4–7days post-fertilization, dpf) from the same wild type (WT) clutch. Individual locomotor activity was tracked at 25 frames per second on a 14hr:10hr light:dark cycle. (B) Ribosome footprint of linc-mipep (also known as lnc-rps25) (top) or linc-wrb (bottom) at 5hours post fertilization (hpf) across annotated transcript length, with putative coding frames in green (+3), orange (+2), or blue (+1); input (control) on bottom tracks. Magenta asterisk marks predicted short open reading frame. RPF, ribosome-protected fragment. (C) Summary of mutagenesis strategy to decode transcript functions. Magenta bars denote CRISPR-targeted area. Mutated/removed sequence is in gray. TSS, transcription start site. sORF, short open reading frame. ATG, start codon. ncRNA, non-coding RNA. Right, phenotypes predicted (check mark) or not predicted (x mark) for each mutant if the gene functions as a regulatory region, noncoding RNA, or protein-coding gene. (D) Stable mutants for linc-mipep: full region deletion (1.78kb deletion, from intron 1 – proximal 3’UTR, top); translation start site deletion that removes the ATG sequence (middle); frameshift deletion (8bp deletion at exon 4, second from bottom); 74bp deletion that removes highly conserved 3’UTR sequence (bottom). (E) Stable frameshift mutant for linc-wrb (11bp deletion, exon 3). (F) Locomotor activity of linc-mipepdel-1.8kb/del-1.8kb;linc-wrbdel-11/del-11 (linc-mipep -/-; linc-wrb -/-, magenta); linc-mipepdel-1.8kb/+;linc-wrbdel-11/+ (linc-mipep +/-; linc-wrb +/-, black); and wild-type (linc-mipep +/+; linc-wrb +/+, blue) sibling-matched larvae over 2 nights. (G) Locomotor activity of wild type (WT, blue) or maternal-zygotic linc-mipepdel1.8kb/del1.8kb;linc-wrbdel11bp/del11bp (linc-mipep;linc-wrb, orange) larvae across two nights. The ribbon represents± SEM. Zeitgeber time is defined from lights ON = 0. (H) Representative daytime locomotor activity tracking of wild type (top 2 rows) and maternal-zygotic linc-mipepdel1.8kb/del1.8kb;linc-wrbdel11bp/del11bp (linc-mipep;linc-wrb, bottom 2 rows) larvae during 1min at 6 dpf. Blue and orange dots represent start and stop locations, respectively.
|