Fig. 3
- ID
- ZDB-FIG-220905-26
- Publication
- Kowatschew et al., 2022 - Spatial organization of olfactory receptor gene choice in the complete V1R-related ORA family of zebrafish
- Other Figures
- All Figure Page
- Back to All Figure Page
|
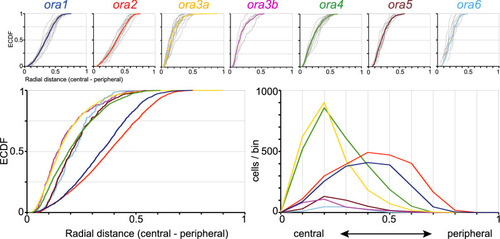
Quantitative analysis of radial position shows three subgroups of ora genes. Radial positions of ora-expressing cells were quantified for all seven ora genes. Complete series of sections from five to fifteen olfactory epithelia were evaluated for each ora gene. Radial position within the section was normalized to maximal radius, i.e., length of the lamella containing the respective labeled cell. Upper row: The resulting distributions of relative radius (from 0, innermost to 1, outermost) are shown unbinned (ECDF). Light grey curves represent the distribution for individual olfactory organs, colored curves represent the cumulative distribution for the respective genes. The color code for the ora genes is the same as in later figures to facilitate comparisons between different positional parameters. Lower row: Overlay of the seven distributions shown individually in upper row, both as ECDF (left panel) and histogram (right panel). Different genes exhibit different centers-of-gravity (mean) and different skewness. Note a segregation in three groups, with ORA3a and ORA3b in a central group, ORA1 and ORA2 in a peripheral group and the remaining three receptors in an intermediate group. |