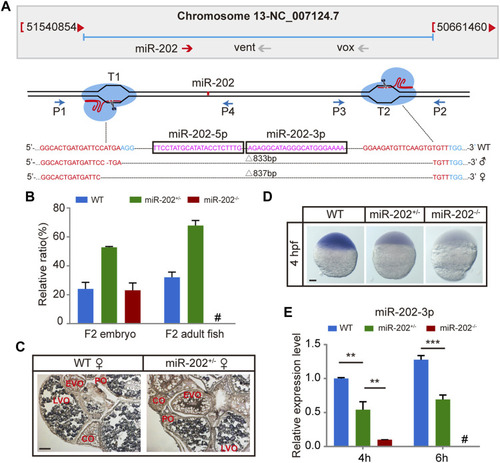
Deletion of miR-202 from the zebrafish genome using CRISPR-Cas9 system. (A) Schematic illustration of CRISPR/Cas9 system used to produce the knock-out lines of miR-202. The miR-202 sequences are in magenta, miR-202-3p and miR-202-5p are marked with a rectangular frame; the sgRNA target sites are in red; the PAM motif (NGG) is shown in blue. The location and direction of primers (P1-P4) used for PCR screening are also shown with arrow. (B) Genotyping of miR-202 mutant F2 embryos at 4hpf and F2 adults (3 month). (C)In situ hybridization of ovaries from miR-202 wild type (WT) and heterozygotes (miR-202+/-) to examine expression of miR-202-3p. The developmental stages of ovarian oocytes include the primary oocytes: perinuclear oocytes (PO) and cortical alveolar oocytes (CO); the mature oocytes: early vitellogenin oocytes (EVO) and late vitellogenin oocytes (LVO). The scale bar is 10 µm. (D) Whole mount in situ hybridization showing expression of miR-202-3p in wild type (WT), heterozygous (miR-202+/-) and homozygous (miR-202−/−) embryos at 4 hpf. The scale bar is 100 µm. (E) qRT-PCR analysis of miR-202-3p expression in wild type, heterozygous and homozygous embryos at 4 hpf and 6 hpf. Error bars, mean ± s.d., n = 3 (biological replicates).
|

