- Title
-
MiR-202-3p determines embryo viability during mid-blastula transition
- Authors
- Hu, R., Xu, Y., Han, B., Chen, Y., Li, W., Guan, G., Hu, P., Zhou, Y., Xu, Q., Chen, L.
- Source
- Full text @ Front Cell Dev Biol
|
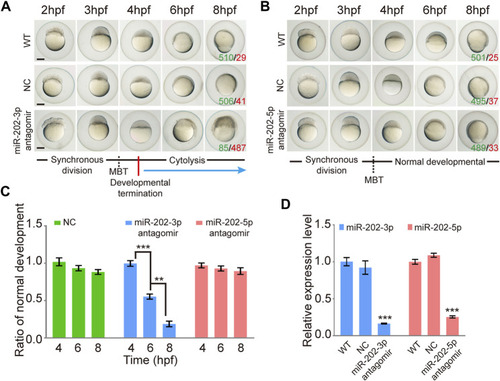
Inhibition of miR-202-3p by an antagomir results in termination of embryonic development at MBT in zebrafish. |
|
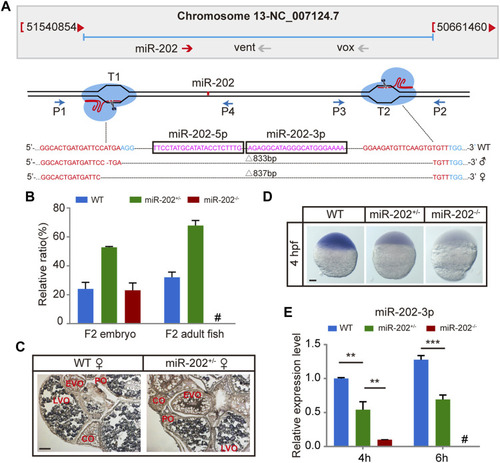
Deletion of miR-202 from the zebrafish genome using CRISPR-Cas9 system. |
|
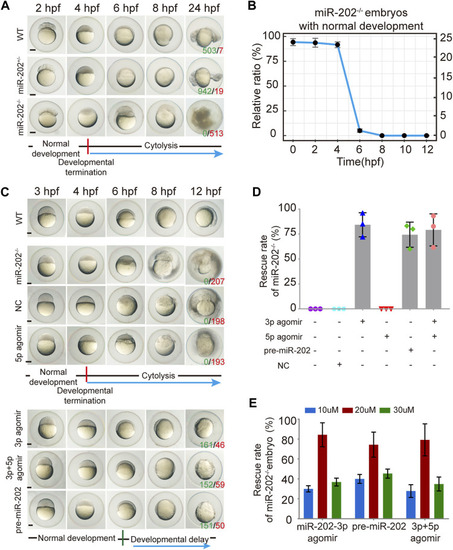
Deletion of the miR-202 locus recapitulated the phenotype of miR-202-3p knockdown. |
|
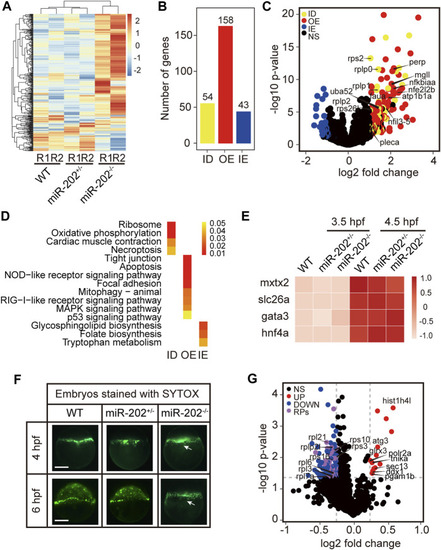
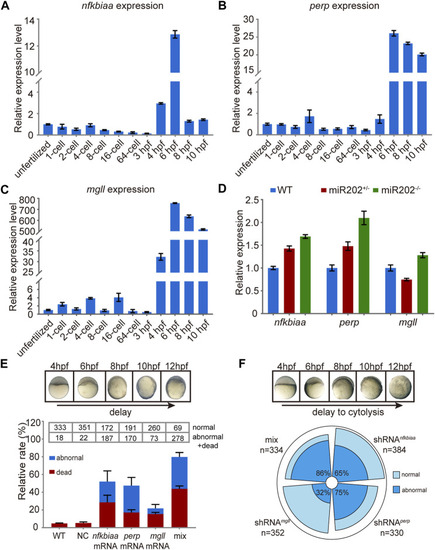
Transcriptomic and proteomic analysis for miR-202 mutant embryos. |
|
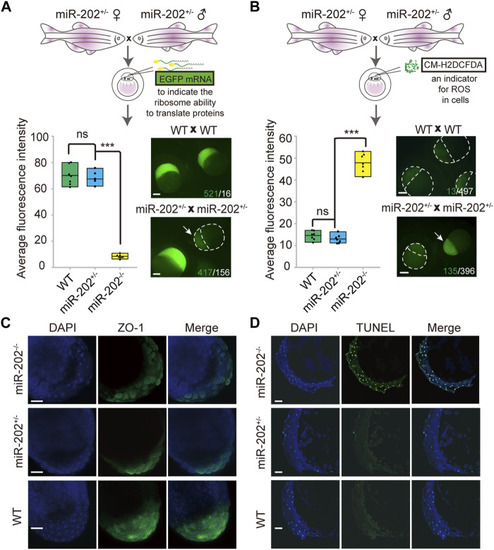
Homeostatic disorders in miR-202−/− embryos. |
|
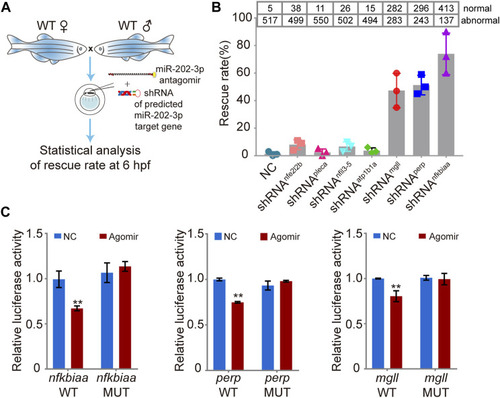
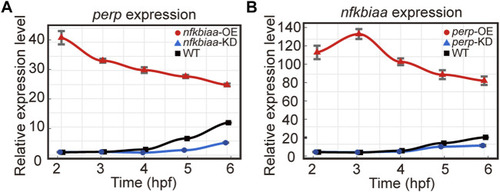
Validation of the function of the miR-202-3p target genes in embryogenesis. |
|
|
|
|
|
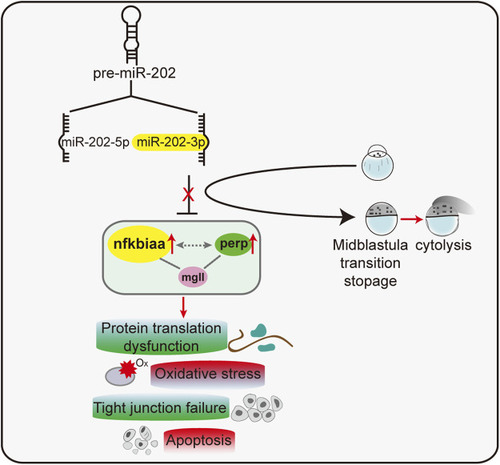
A proposed model of a miR-202-3p mediated regulatory network that determines embryonic viability during MBT in zebrafish. |