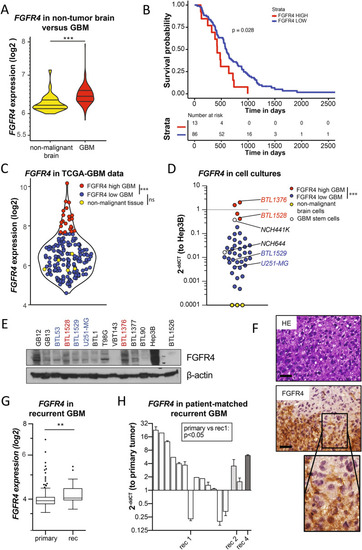
FGFR4 is overexpressed in a highly aggressive GBM subgroup, associated with tumor recurrence. AFGFR4 mRNA levels in non-tumor brain (n = 28) and GBM (n = 219) in the REMBRANDT dataset are shown as violin plots. B Kaplan Meier survival analysis of GBM patients from the REMBRANDT dataset stratified for FGFR4 expression (n = 99.) CFGFR4 mRNA levels in non-malignant brain (yellow, n = 5), low- (blue, n = 147), and high- (red, n = 22) expressing GBM subgroups of the TCGA-GBM RNA sequencing data. DFGFR4 mRNA levels detected by qRT-PCR in GBM (n = 40), glioblastoma stem cells (n = 2, white) models, and non-malignant brain tissue extracts (n = 3, yellow) are shown relatively to Hep3B positive control (2−ddCT). FGFR4high and FGFR4low GBM models selected for further analyses are highlighted, respectively. E Detection of FGFR4 (several bands due to increasing glycosylation) in membrane-enriched fractions from selected GBM cell models and Hep3B by Western blot, with β-actin as loading control. F FGFR4 IHC staining of BTL1376 tumor material is opposed to haematoxilin eosin (HE) stain. Scale bars: 50 µm. GFGFR4 mRNA data (TCGA-GBM-HG-U133A) of primary (n = 497) and recurrent (n = 16) GBM are visualized. HFGFR4 mRNA levels in patient-matched primary GBM (n = 14) and sequential recurrences (rec1, n = 13; rec2, n = 2; rec4, n = 1). Statistical analyses: log-rank test B; Wilcoxon test C; maximization of the t-statistics D; Student’s t-tests A,G,H;. n.s. = not significant, *p < 0.05, **p < 0.01, ***p < 0.001
|

