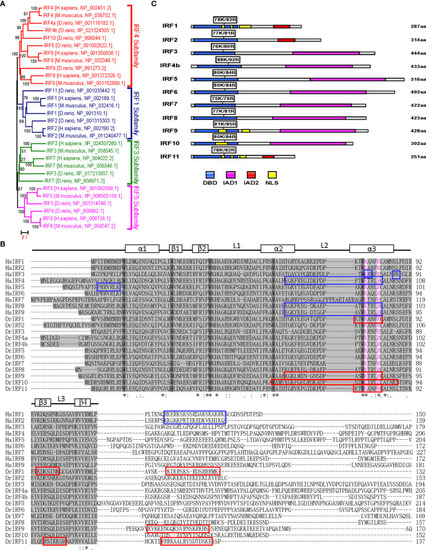
Figure 5
|
Figure 5 K78 and R82 of α3 helices in DBD domain of IRF11 are conserved across IRF family members. (A) Phylogenetic tree analysis of zebrafish and human IRF family members showing four IRF subfamilies. The phylogenetic tree was constructed by a neighbor-joining method in MEGA 5.0. The bootstrap confidence values shown at the nodes are based on 1000 bootstrap replications. (B) Multiple alignments of zebrafish IRF proteins showing a highly conserved DBD and the distribution of NLS motifs identified previously. DBD domains are gray with five conserved tryptophans. The symbols, including α-helices, β-strands and loops, indicates the secondary structures of DBD, which are marked by rectangle and line. Based on publications, the identified NLS motifs of zebrafish IRF1/10/11 are indicated by red boxes, and the NLS motifs of human IRF1/2/3/4/5/8/9 in blue boxes. Zebrafish IRF11 has a tripartite NLS motif composed of NLS1, NLS2 and NLS3, with the conserved K78 and R82 that are highlighted in purple. Identical (*) and similar (: or.) amino acid residues are indicated. (C) Schematic diagram of zebrafish IRF proteins, showing the position of two conserved basic residues corresponding to K78 and R82 of zebrafish IRF11. All mutants of zebrafish IRF protein were generated by combined mutation of the corresponding two residues to alanine. |