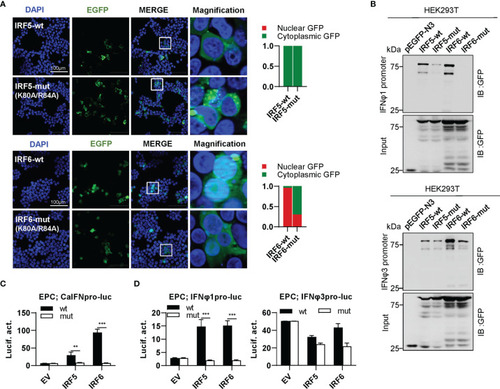
The two basic residues are essential for IRF5/6 to regulate IFN response. (A) Mutation of the two residues impaired the constitutively nuclear accumulation of IRF6 but not of IRF5. Left panels: HEK293T cells seeded overnight on microscope slide cover-glasses in six-well plates were transiently transfected as in Figure 4, with indicated plasmids (2μg each) for 24h, followed by confocal microscopy examination. The last column showed magnification view of the area highlighted in the box. Right panels: The intensities of nucleus/cytoplasm GFP were quantitated using the ImageJ processing program and normalized to that of the empty construct pEGFP-N3, which was set to 1:1. (B) Pull-down analysis of the binding affinity of wild types and mutants of IRF5/6 to IFNφ1/IFNφ3 promoters. DNA pull-down assays were performed as in Figure 3B, by incubating biotin-labeled IFNφ1 or IFNφ3 promoter DNA with appropriate amounts of HEK293T cell lysates, where cells were transfected for 30h with wild types or mutants of IRF5/6, respectively. (C, D). IRF5/6 mutants failed to stimulate the activation of crucian carp IFN promoter (C), and zebrafish IFNφ1/IFNφ3 promoters (D). EPC cells seeded in 48-well plates overnight were transfected with CaIFNpro-luc (C), IFNφ1pro-luc or IFNφ3pro-luc (D), together with the indicated IRF plasmids (100ng each). 30h later, cells were harvested for luciferase assays. (***P < 0.001; **P < 0.01).
|

