Figure 4
- ID
- ZDB-FIG-220203-52
- Publication
- Zhu et al., 2021 - Nucleolar GTPase Bms1 displaces Ttf1 from RFB-sites to balance progression of rDNA transcription and replication
- Other Figures
- All Figure Page
- Back to All Figure Page
|
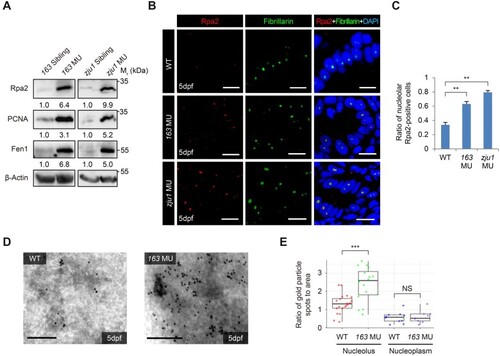
Bms1l mutations cause replication fork stall. (A) Western blotting showing a drastic increase of Rpa2, PCNA, and Fen1 protein levels in 5dpf bms1lsq163/sq163 and bms1lzju1/zju1 mutants compared with their siblings. β-Actin: loading control. (B and C) Coimmunostaining of Rpa2 and Fibrillarin in gut epithelia was performed (B). The ratio of the nucleolar Rpa2-positive cells was significantly higher in bms1lsq163/sq163 and bms1lzju1/zju1 mutants than in WT at 5dpf (C). Samples: two WT embryos, 544 cells examined; three bms1l163/sq163 embryos, 845 cells examined; two bms1lzju1/zju1 embryos, 277 cells examined. Scale bar, 10 μm. (D and E) Representative images showing Immuno-TEM of Rpa2 in hepatocytes dissected from the WT and bms1lsq163/sq163 embryos at 5dpf (D). Statistical analysis showing a significant accumulation of Rpa2-positive gold particles in the nucleoli but not in the nucleoplasm in bms1lsq163/sq163 hepatocytes compared with WT (E). For each genotype, 17‒20 nucleoli were examined. Each dot represents the ratio of gold particle numbers against area of the nucleolus or nucleoplasm in one cell. Scale bar, 0.2 μm. In C and E, **P < 0.01, ***P < 0.001; NS, no significance. |