FIGURE 1
- ID
- ZDB-FIG-220131-265
- Publication
- Zhang et al., 2022 - Conservation of Differential Animal MicroRNA Processing by Drosha and Dicer
- Other Figures
- All Figure Page
- Back to All Figure Page
|
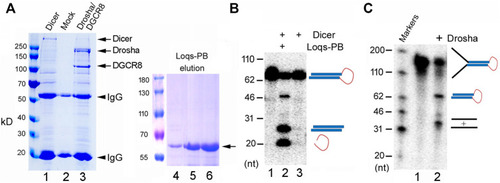
Purification and activity assays of the fruitfly miRNA processing enzymes. (A) Purification of recombinant Dicer, Drosha/DGCR8 from transfected 293T cells (lanes 1–3) and Loqs-PB from overexpressing bacteria (lanes 4–6). 293T cells were transfectd with plasmids expressing the FLAG-tagged Dicer or FLAG-tagged Drosha and Myc-tagged DGCR8. Following immunoprecipitation with an anti-FLAG antibody, proteins were detected by gel electrophoresis and Coomassie staining. Lane 1: The immunoprecipitate of FLAG-Dicer; lane 2: immunoprecipitate from mock-transfected cells; lane 3: immunoprecipitate of FLAG-Drosha and Myc-DGCR8; lanes 4–6: different elution fractions of His-tagged Loqs-PB after nickle beads purification. Arrows points to the expected fruitfly proteins, and arrowheads the IgG heavy and light chains. Protein markers (in kilodaltons, kD) are indicated on the left. (B) Dicer activity assay. The 32P-labeled dme-pre-miR-375 was incubated with Dicer alone or together with Loqs-PB at 37°C for 30 min, fractionated on a 12% denaturing gel, and analyzed by phosphorimaging. DNA markers with size of the individual bands in nucleotide (nt) are indicated on the left, and schematics of the substrate and cleavage products shown on the right. (C) Drosha processing assay. The 32P-labeled dme-pri-let-7 substrate was incubated with Drosha/DCGR8 (Drosha in short) at 37°C for 60 min. Samples were fractionated on a 10% denaturing gel and analyzed by phosphorimaging. Labels are the same as in (B). |