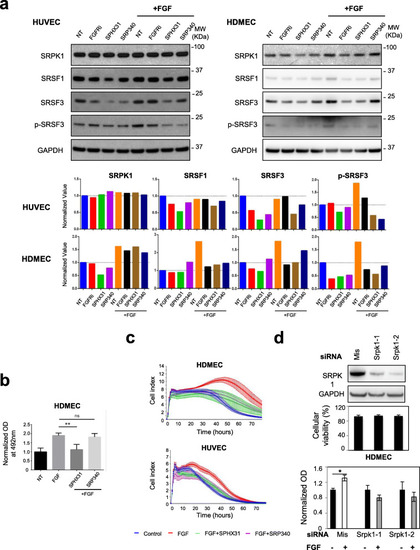
FGF2-mediated endothelial cells proliferation and survival requires SRPK1 activity. aUpper panel: immunoblots of the indicated proteins in HUVEC (left) and HDMEC (right) treated or not (NT) for 72 h with 10nM AZD4547 (FGFRi), 5μM SPHINX31 (SPHX31), or 10μM SRPIN340 (SRP340) in the presence or absence of 3nM FGF-2 as indicated. GAPDH was used as a loading control. Data representative of 2 (HUVEC) and 3 (HDMEC) independent experiments are presented. Lower panel: semi-quantification using ImageJ software of the signals obtained for the indicated proteins relative to GAPDH signal. The ratio obtained for the NT group was arbitrarily assigned the value 1. b MTS test was used to quantify HDMEC cellular viability in response to 72 h treatment with or without 3nM FGF-2 in the presence or absence of 5μM SPHINX31 (SPHX31) or 10μM SRPIN340 (SRP340) (n=4 technical replicates, unpaired t test, **p<0.01, ns: not significant). c xCELLigence assay was used to assess HUVEC and HDMEC cell adhesion, proliferation, and viability in response to 3nM FGF-2 with or without 5μM SPHINX31 (SPX31) or 10μM SRPIN340 (SRP340) (n=3). d HDMEC was transfected for 48 h with either mismatch (mis) or distinct SRPK1 siRNAs as indicated. Upper panel: representative immunoblot of SRPK1 knockdown. GAPDH was used as a loading control (n=3). Middle panel: cellular viability (%) was assessed after trypan blue staining in transfected HDMEC just before plating (n=5). Lower panel: MTS assay was used to quantify cellular viability in HDMEC deprived of SRPK1 (Srpk1) or not (Mis) and treated (+) or not (−) with 3nM FGF-2 for 48 h. Mean ± SD (n=3; unpaired t test, *p<0.05)
|

