Figure 1
- ID
- ZDB-FIG-200421-72
- Publication
- Giribaldi et al., 2020 - Synthesis, Pharmacological and Structural Characterization of Novel Conopressins from Conus miliaris
- Other Figures
- All Figure Page
- Back to All Figure Page
|
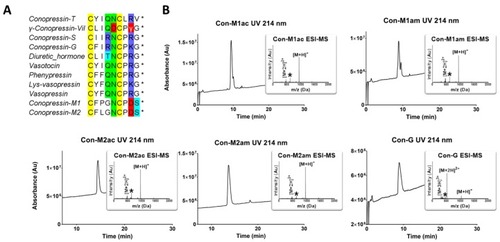
RP-HPLC/ESI-MS analyses of the synthesized conopressins and alignment of conopressin-related sequences. ( |