|
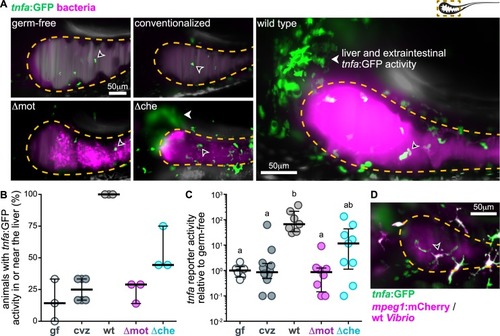
Motile bacterial cells induce local and systemic <italic>tnfa</italic> expression.(A) Maximum intensity projections acquired by LSFM of the foregut region of tnfa:GFP transgenic zebrafish raised germ-free, with a complex microbial community (conventionalized), or colonized solely with dTomato-expressing (magenta) wild-type Vibrio, Δmot, or Δche. Animals were imaged at 24 hpi. Dashed lines mark the approximate intestinal boundaries. Empty arrowheads mark host cells with tnfa:GFP reporter activity. Solid arrowheads mark tnfa:GFP reporter activity in extraintestinal tissues in or near the liver. (B) Percent of zebrafish subjected to different colonization regimes with tnfa:GFP activity in or near the liver; >6 animals/group were blindly scored by 3 researchers. Bars denote medians and interquartile ranges. (C) Total GFP fluorescence intensity across the foregut region normalized to median gf fluorescence intensity. Bars denote medians and interquartile ranges. Letters denote significant differences. p < 0.05, Kruskal-Wallis and Dunn’s multiple comparisons test. (D) Maximum intensity projections acquired by LSFM of the foregut region of a tnfa:GFP, mpeg1:mCherry (magenta) transgenic zebrafish colonized with dTomato-expressing wild-type Vibrio (magenta). Animal was imaged at 24 hpi. Open arrowhead indicates a tnfa+/mpeg1+ cell. Underlying data plotted in panels B and C are provided in S1 Data. cvz, conventionalized; gf, germ-free; GFP, green fluorescent protein; hpi, hours post inoculation; LSFM, light sheet fluorescence microscopy.
|