Fig. 2
- ID
- ZDB-FIG-200306-11
- Publication
- Lee et al., 2020 - Regenerating zebrafish fin epigenome is characterized by stable lineage-specific DNA methylation and dynamic chromatin accessibility
- Other Figures
- All Figure Page
- Back to All Figure Page
|
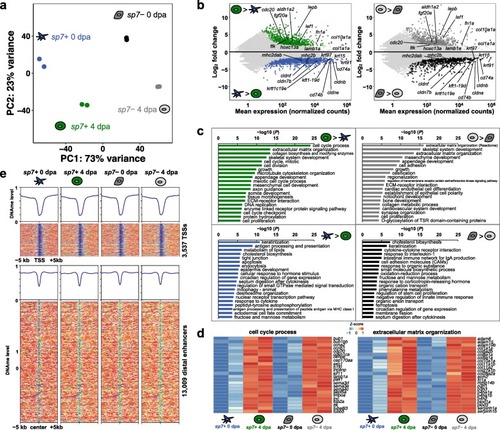
Regeneration-specific genes are activated independent of DNA methylation changes. |