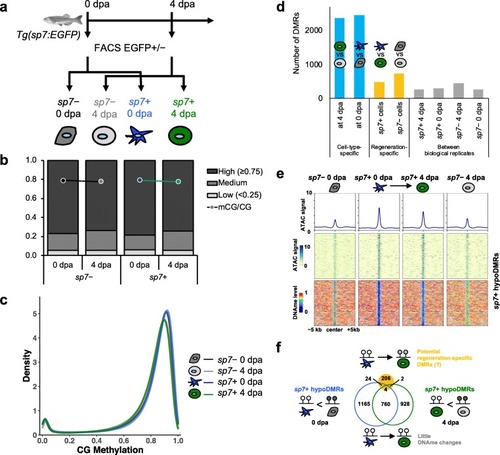
Lineage-specific DNA methylation signatures are stably maintained during fin regeneration. a Experimental scheme of sorting sp7+ and sp7− cells from uninjured and regenerating zebrafish fin by using FACS. b Global CpG methylation levels (mCG/CG) and fraction of total CpGs with low (< 25%), medium (≥ 25% and < 75%), and high (≥ 75%) methylation levels of sp7+ and sp7− cells during zebrafish fin regeneration. c Distribution of genome-wide CpG methylation levels of each cell type. Bimodal distribution of two CpG populations at high and low methylation levels is observed. d Number of DMRs identified between two biological replicates (gray bars), between two different time points in the same cell type (regeneration-specific, yellow bars) or between two different cell types at the same time point (cell-type-specific, blue bars). e ATAC-seq signals (top) and DNA methylation levels (bottom) over 10-kb regions centered on a total of 2883 sp7+ cell-specific hypoDMRs. Average ATAC-seq signals were plotted on top of each heatmap (line plots). f Venn diagram of sp7+ cell-specific hypoDMRs (blue and green circles for 0 dpa and 4 dpa, respectively) intersecting with potential regeneration-specific DMRs in sp7+ cells (yellow filled circle). Only 30 (1.0%) of sp7+ cell-specific hypoDMRs were predicted as potential regeneration-specific DMRs
|

