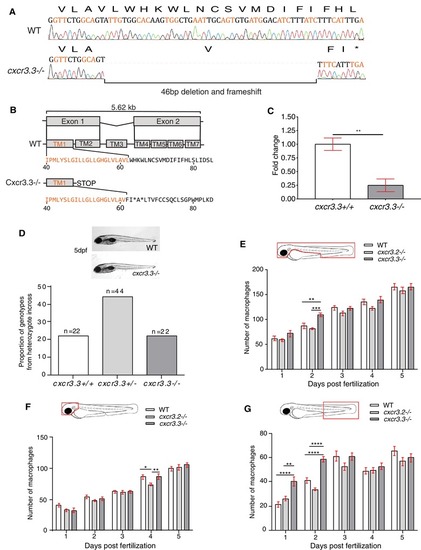
cxcr3.3 mutant larvae do not show morphological aberrations or major differences in macrophage development. A 46 bp deletion was induced in the cxcr3.3 gene using CRISPR‐Cas9 technology (A). The deletion is located in the first exon (orange), at the very end of the first TM domain (TM1).The mutation shifts the reading frame and results in a premature stop codon (B). Nonsense‐mediated decay assessment suggests that the cxcr3.3 mutant gene codes a truncated Cxcr3.3 protein (C). No evident morphological aberrations were observed in cxcr3.3–/– larvae within the first 5 dpf and the mutant allele segregated following Mendelian proportions (D). Macrophage development was faster in cxcr3.3–/– embryos at 2 dpf but reverted to WT and cxcr3.2–/– pace after day 3 (E). Fewer macrophages were found in the head area of cxcr3.2–/– larvae only at day 4 (F), whereas there were more macrophages in the tail region in cxcr3.3–/– (G). The cell numbers corresponding to each day are the average of 35 larvae of each of the 3 groups (genotypes). Data were analyzed using a two‐way ANOVA and are shown as mean ± sem (ns P > 0.05, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001)
|

