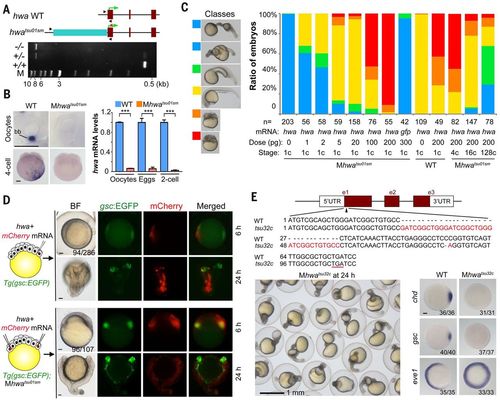
Identification and verification of tsu01sm mutant gene. (A) Top: Illustration of the zebrafish hwa wild-type and tsu01sm mutant alleles. Arrowheads indicate the positions of PCR primers. The green arrow indicates the translation start site. Bottom: Electrophoretic result of PCR products in wild-type, heterozygous, and homozygous mutant embryos. M, molecular weight markers. (B) hwa mRNA detection by WISH (left) and by qRT-PCR (right). bb, Balbiani body in stage I oocyte. For qRT-PCR, oocytes are a pool of stage I–III oocytes and eggs are squeezed from females. The relative hwa mRNA levels are averages (±SEM) from three independent experiments, normalized to eif4g2a levels. ***P < 0.001. Scale bars, 100 μm. (C) Mhwatsu01sm mutant embryos form the body axis or are dorsalized by hwa overexpression. Left: Morphology of classified embryos at 24 hpf. Right: Ratios of embryos in different classes. (D) Induction of secondary body axis by ectopic hwa mRNA overexpression. One blastomere of 32-cell stage Tg(gsc:EGFP) transgenic embryos or two opposite blastomeres of 32-cell stage Tg(gsc:EGFP);Mhwatsu01sm mutant embryos were injected with 50 pg of hwa and 50 pg of mCherry mRNAs, as illustrated at left. Embryos were observed dorsally (top) or laterally (bottom) at 6 hpf under a fluorescence dissection microscope, and those with two dorsal organizers (EGFP-positive) were observed again at 24 hpf. The ratio of embryos with double organizers is indicated. Scale bars, 100 μm. (E) Generation of a hwa mutant allele by Cas9 knockout. Top: Illustration of the mutant allele with an underlined premature stop codon. Bottom left: A group of Mhwatsu32c mutant embryos at 24 hpf. Bottom right: Alteration of the dorsal markers gsc and chd and the ventral marker eve1 at shield stage; scale bar, 100 μm.
|

