Fig. 3
- ID
- ZDB-FIG-180710-47
- Publication
- Kenyon et al., 2018 - Generation of a double binary transgenic zebrafish model to study myeloid gene regulation in response to oncogene activation in melanocytes
- Other Figures
- All Figure Page
- Back to All Figure Page
|
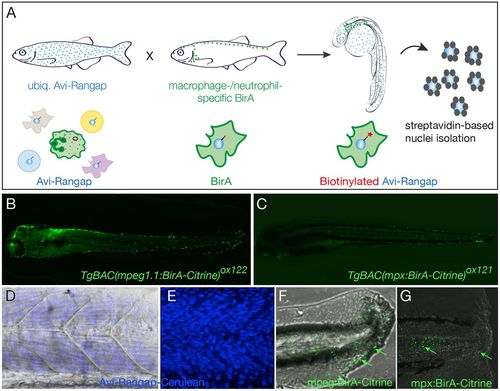
Binary transgenic zebrafish model for regulatory profiling of myeloid cells. (A) Schematic of myeloid nuclear biotagging system. When biotagging effector transgenic zebrafish line ubiquitously expressing Avi-tagged Rangap for biotinylation of nuclei is crossed to biotagging driver line expressing BirA in myeloid-specific manner, only the macrophage or neutrophil nuclei will be biotinylated. (B) Transgene expression in macrophage-specific BirA driver, TgBAC(mpeg1:BirA-Citrine)ox122, is amplified with anti-GFP–Alexa-Fluor-488 to show expression in macrophages. (C) TgBAC(mpx:BirA-Citrine)ox121 biotagging transgenic driver shows neutrophil-specific expression. (D,E) Projection of confocal microscope images of Tg(bactin:Avi-Cerulean-Rangap)ct700a shows ubiquitous expression of the biotag effector specifically on nuclei, across somite region of the embryo, with (D) and without (E) bright field background. The images in D and E are taken in different embryos. (F,G) Tailfins of transgenic fish transected at 3 dpf show responding macrophages in mpeg1:BirA-Citrine (F) and neutrophils in mpx:BirA-Citrine (G) larvae at 5 hpi or 1 hpi, respectively, as indicated by green arrows. |
| Genes: | |
|---|---|
| Fish: | |
| Condition: | |
| Anatomical Terms: | |
| Stage: | Day 5 |