Fig. 5
|
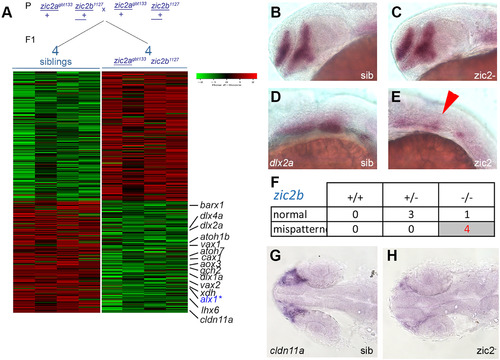
RNA sequencing transcriptome analysis identifies a set of Zic2-dependent targets. A: Embryos derived from zic2agbt133/+; zic2b1127/+ parents were sorted by presence or absence of coloboma into zic2 mutants and sibling groups, respectively. RNA extracted from individual embryos (4 wildtype and 5 with coloboma) was used to prepare cDNA libraries for illumina high-throughput sequencing. Genes with assigned value of False-Discovery Rate below 0.05 were preliminarily selected. The heat-map color represents relative expression levels of differentially expressed genes; 4 out of 5 coloboma-representing libraries are shown to maintain visual balance with the 4 normal sibling samples. B, C: Representative sibling and zic2 mutant embryos derived from zic2agbt133/+;zic2buw1116/+ parents show normal dlx2a expression by WISH in the telencephalon and diencephalon. D: normal dlx2a expression in branchial arch primordia. E: depleted branchial arch dlx2a expression (arrowhead) was observed in 7 out of 127 (6%, 3 expts.) of embryos from this cross. F: Only zic2b- homozygotes exhibited dlx2a reduction in branchial arch primordia. G: normal cldn11a expression adjacent to the optic stalk of embryo with normal retinal morphology. H: depleted cldn11a expression in zic2 mutant with coloboma. Embryos in B – E are shown in lateral views, anterior to the left. Embryos in G and H are shown in dorsal views, anterior to the left. |
Reprinted from Developmental Biology, 429(1), Sedykh, I., Yoon, B., Roberson, L., Moskvin, O., Dewey, C.N., Grinblat, Y., Zebrafish zic2 controls formation of periocular neural crest and choroid fissure morphogenesis, 92-104, Copyright (2017) with permission from Elsevier. Full text @ Dev. Biol.