Fig. S2
- ID
- ZDB-FIG-101223-11
- Publication
- Diz-Munoz et al., 2010 - Control of Directed Cell Migration In Vivo by Membrane-to-Cortex Attachment
- Other Figures
- All Figure Page
- Back to All Figure Page
|
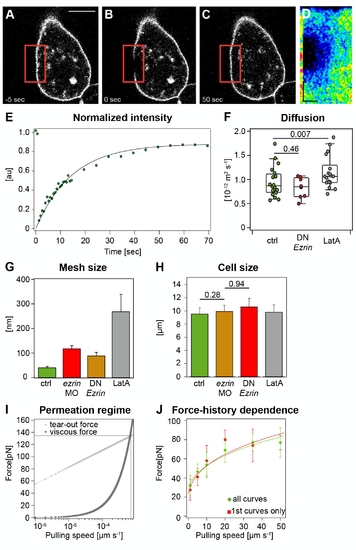
Physical properties of the plasma membrane and cortex in control and ERM-deficient prechordal plate progenitor cells. (A–F) Plasma membrane fluidity measurements of control and ERM-deficient prechordal plate progenitor cells. Sequential images of a typical FRAP experiment before bleaching (A), directly after bleaching (B), and after complete recovery (C). Red square demarcates bleached region. (D) Kymograph of the bleached region. The kymograph was calculated on a line (width = one pixel) encompassing the bleached region of the cell membrane within the red box. Recovery occurs from the rims of the bleached area. Scale bars: in y = 5 μm and in x = 9 s. (E) Example of a recovery curve for a prechordal plate progenitor cell expressing GAP43-GFP. (F) Diffusion coefficient extracted from FRAP experiments in control, ERM-deficient, and LatA-treated prechordal plate progenitor cells. For more details see Text S1. (G) Average lateral separation between plasma-membrane-to-cortex cross-linking molecules of control, ERM-deficient, and LatA-treated prechordal plate progenitor cells. Error bars indicate error propagated from the fit. (H) Cell radius measured during cortical tension measurements. Error bars indicate error propagated from the fit. p-value was estimated using t test. (I) Estimation of the range of velocities for which Equation 2 is valid. The plot represents theoretical curves, computed as in [52], of the forces exerted on transmembrane proteins during tether extraction. The friction force (Ff, dark grey squares) due to the flow of membrane into the tether increases linearly with pulling velocities, whereas the rupture force (Fr, light grey squares), at which the transmembrane protein would unbind from the cortex, increases logarithmically. The intersection between these two curves (600 μm·s-1) gives the critical velocity at which transmembrane proteins unbind from the cortex. Therefore, the velocities we used for tether pulling (lower than 100 μm·s-1) are suitable to estimate ν. The parameters used to compute the curves are the membrane viscosity (1.5x10-7 Pa•m•s; see Materials and Methods) and the typical distance between cortex–membrane linkers (0.35 nm [53]). (J) Comparison between the extrusion force of the first tether that has been extruded from a cell and the tether force of all measurements shows that force is not influenced by the pulling history. The fit parameters are a = 9,969 and F0 = 32 pN for “all curves” and a = 11,612 and F0 = 25 pN for “first curves.” |