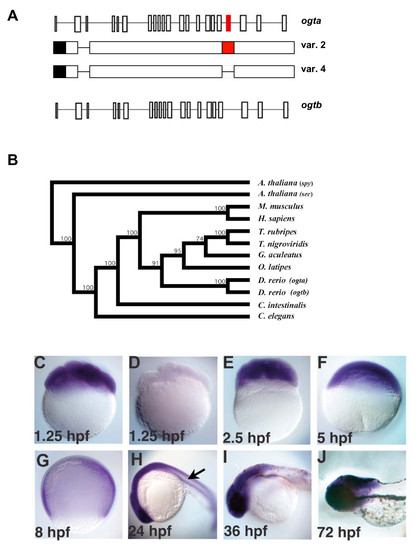
The origin and expression pattern of zebrafish ogt genes. (A) Schematic diagram of zebrafish ogta and ogtb loci (formerly known as copy I and copy II, respectively), which are located on Chromosome 14 [46]. Diagrams of variant 2 and 4 transcripts from the ogta gene are represented under the diagram of the ogta locus. The black box represents the 3′ UTR, while the red boxes in the ogta locus and variant 2 transcript represent the alternatively spliced exon 19, which has been proposed to inhibit O-GlcNAc Transferase activity [46]. Exon 19 is absent from variant 4 transcripts. (B) Phylogenetic analysis of ogt genes constructed using the neighbor-joining method using aligned amino acid sequences (see materials and methods for gene identifier numbers for each of the sequences used). The A. thaliana ogt gene spy was used to root the tree, since it is the most divergent ogt gene we analyzed. Bootstrap values for 1,000 repetitions are indicated at the nodes. (C-J) A time course of ogt gene expression from 8-cell (1.25 hpf) (C) to 72 hpf (J) of development reveals that ogt is dynamically expressed in early embryos. ogt is ubiquitously expressed in cleavage stage (C, E), blastula stage (F) and gastrula stage (G) embryos. Signal is not detected in embryos incubated with sense probe (D). At 24 hpf, ogt transcripts are expressed at high levels in the brain and lower levels trunk and tail (H). At 36 hpf (I) and 72 hpf (J), ogt transcripts are restricted to the brain.
|

