Fig. S1
- ID
- ZDB-FIG-090304-4
- Publication
- Slanchev et al., 2009 - Control of Dead end localization and activity – Implications for the function of the protein in antagonizing miRNA function
- Other Figures
- All Figure Page
- Back to All Figure Page
|
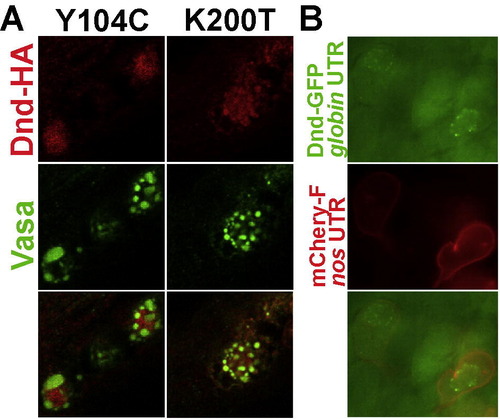
Sub-cellular localization of Dnd versions in PGCs and somatic cells. (A) Similar to other loss of function mutations within the RRM (see Fig. 5), the Y104C mutated Dnd is localized to the nucleus. In contrast, Dnd with loss of function mutation K200T, which resides outside the RNA binding domain, was localized to the germ-cell granules. (B) Over-expression of Dead end in somatic cells using globin 3′UTR. Within the PGCs, outlined in red using membrane bound mCherry under the control of nanos1 3′UTR, Dnd is localized to the germ-cell granules. In the somatic cells, Dnd is ubiquitously distributed within the cell. |
Reprinted from Mechanisms of Development, 126(3-4), Slanchev, K., Stebler, J., Goudarzi, M., Cojocaru, V., Weidinger, G., and Raz, E., Control of Dead end localization and activity – Implications for the function of the protein in antagonizing miRNA function, 270-277, Copyright (2009) with permission from Elsevier. Full text @ Mech. Dev.