Person
Plessy, Charles
|
|
Biography and Research Interest
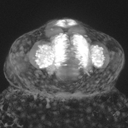
Image : Frontal view of a 24h -8.4ngn1:GFP transgenic fish. Nice view on the telencephalon (medial), and on the olfactory bulb (lateral). The dorsalmost structure is the pineal body.
Ph.D. Research Topic: Integration of spatial information by neurogenin1 in zebrafish neural plate.
Ph.D. Research Topic: Integration of spatial information by neurogenin1 in zebrafish neural plate.
Current Research Topic: Promotome profiling of large collections of single cells for systems biology.
