- Title
-
A maternal-to-zygotic-transition gene block on the zebrafish sex chromosome
- Authors
- Wilson, C.A., Postlethwait, J.H.
- Source
- Full text @ G3 (Bethesda)
|
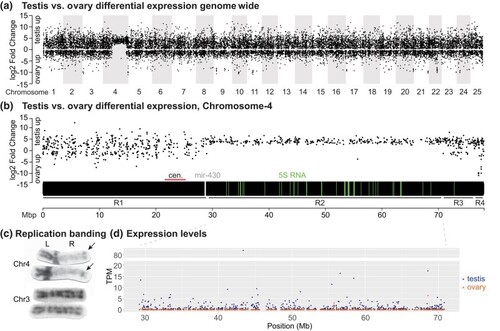
Testis vs ovary differential gene expression. a) Testis vs ovary differential expression plotted against genomic position across the 25 zebrafish chromosomes. The plot shows unique and specific upregulation of testis genes vs ovary genes on Chr4, the sex chromosome. b) Testis vs ovary differential expression across Chr4. The plot identified 4 distinct regions: R1, with values similar to the bulk of the genome; R2, with testis expression on average much higher than ovary expression; R3, again with values similar to most of the genome; and R4, with some strongly ovary-biased genes and coincident with |
|
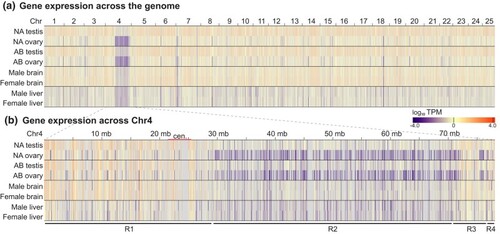
Gene expression in gonads and in somatic organs measured by log10 TPM. a) A heatmap showing TPM levels for annotated protein-coding genes across the genome in male and female gonads of strain NA and AB and in male and female somatic organs (brain and liver) for strain AB plotted against position. Color scale below at right. b) A heatmap showing TPM along the length of Chr4 for the same samples as in a). Plots revealed low transcript levels in Region-2, especially in ovaries of both Nadia and AB, and that, although gene expression was also low in Region-2 in strain AB brain and liver, it was not sexually dimorphic in these somatic organs. |
|
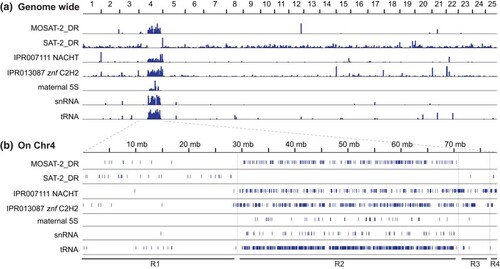
Distinctive features of Chr4. a) Distribution of sequences across the 25 zebrafish chromosomes for satellite sequences MOSAT-2 and SAT2, genes encoding the protein-coding domains IPR007111 NACHT, genes encoding IPR013087 C2H2-type zinc fingers, and certain nonprotein-coding genes including maternal 5S rRNA genes (somatic 5S rRNA genes on Chr18 not shown), snRNA genes, and tRNA genes. b) Positions of these sequences along Chr4 are specifically enriched or excluded from Region-2 ( |
|
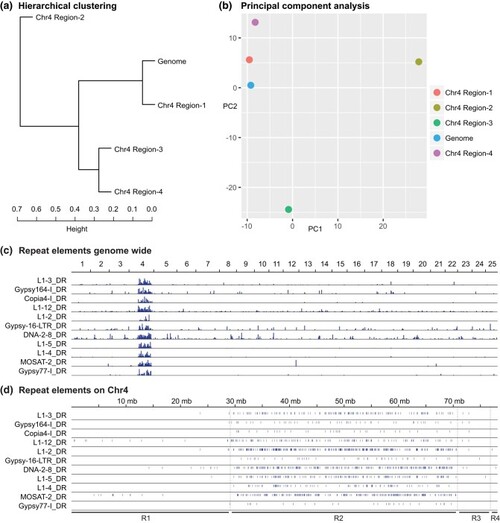
Distribution of repetitive elements. a) Hierarchical clustering analysis of repetitive element composition in the 4 regions of Chr4 and in the rest of the zebrafish genome (Chr1–Chr3 plus Chr5–Chr25). b) PCA analysis of repetitive element composition shows that Region-2 of Chr4 is distinct from the rest of the genome. c) Genomic distribution of several repeat elements contributing to PC1 shows them to be clustered on Chr4R. d) Distribution of these repeats along Chr4 shows that they tend to concentrate in Region-2. |
|
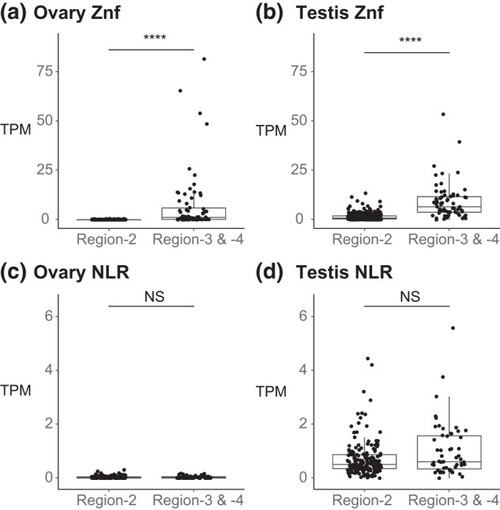
Comparison of expression levels of zinc finger and NLR genes in Region-2 vs Region-3 + Region-4. a) In the ovary, |
|
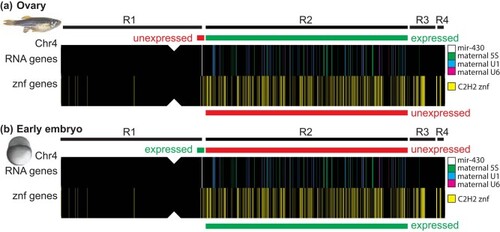
A maternal-to-zygotic-transition gene block. a) In the ovary, oocytes express maternal 5S rRNA (green vertical bars on the chromosome sketch) and spliceosome genes (U1, U6; blue and mauve) in Region-2 but not |