- Title
-
Comparative transcriptomics coupled to developmental grading via transgenic zebrafish reporter strains identifies conserved features in neutrophil maturation
- Authors
- Kirchberger, S., Shoeb, M.R., Lazic, D., Wenninger-Weinzierl, A., Fischer, K., Shaw, L.E., Nogueira, F., Rifatbegovic, F., Bozsaky, E., Ladenstein, R., Bodenmiller, B., Lion, T., Traver, D., Farlik, M., Schöfer, C., Taschner-Mandl, S., Halbritter, F., Distel, M.
- Source
- Full text @ Nat. Commun.
|
mmp9:Citrine identifies mature neutrophils. |
|
Mmp9+ neutrophils show functions of mature neutrophils. |
|
scRNA-seq of zebrafish neutrophils reveals a continuous maturation process. |
|
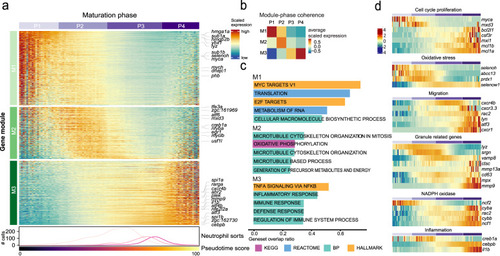
Trajectory analysis uncovers the underlying cell phases and governing gene modules. |
|
Cebpb expression is associated with neutrophil maturation in zebrafish. |
|
Cebpb regulates aspects of late neutrophil maturation in zebrafish. |
|
Alignment of expression trajectories across zebrafish, mouse and human reveals concordant and divergent stages of maturation. |
|
A pan-species neutrophil maturation signature shows the enrichment of mature neutrophils in BM of metastatic neuroblastoma patients. |
|
Model for neutrophil development in zebrafish based on scRNA sequencing. Association of gene expression with modules and phases as determined by scRNA-seq. Granule stage and morphology were assumed by comparing gene expression (as in Fig. |