- Title
-
Bmp4 in Zebrafish Enhances Antiviral Innate Immunity through p38 MAPK (Mitogen-Activated Protein Kinases) Pathway
- Authors
- Chen, L., Zhong, S., Wang, Y., Wang, X., Liu, Z., Hu, G.
- Source
- Full text @ Int. J. Mol. Sci.
|
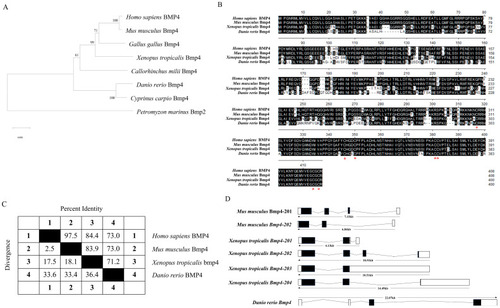
Bioinformatic analysis of |
|
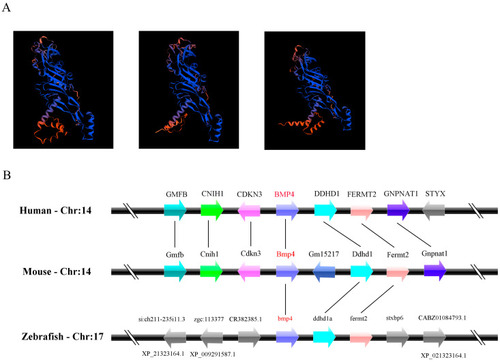
Comparison of the 3-D structure of BMP4 proteins and the loci of |
|
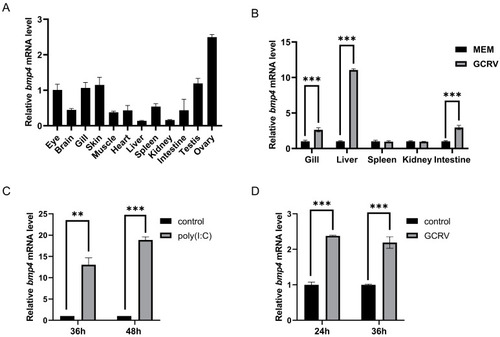
The |
|
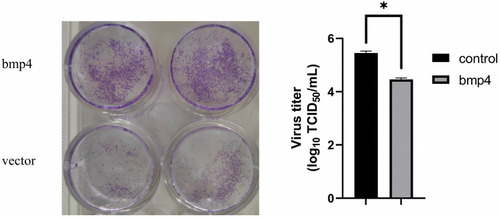
Antiviral function of Bmp4 in vitro. Overexpressing |
|
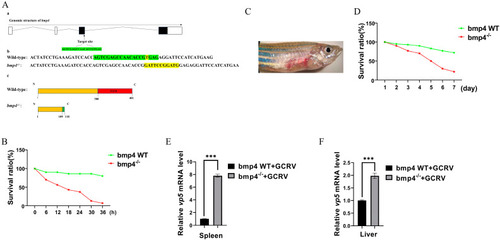
Antiviral function of Bmp4 in vivo. ( |
|
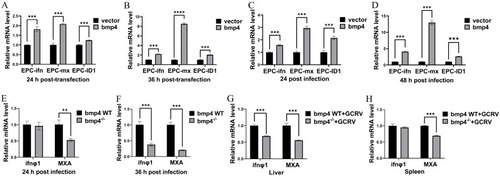
Bmp4 promotes the expression of antiviral genes. ( |
|
KEGG analysis of the pathways through enrichment of DEGs between |
|
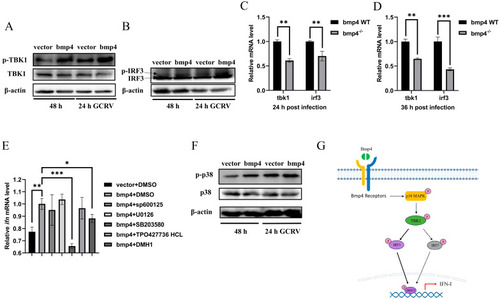
Bmp4 increases Tbk1-Irf3 antiviral signaling via p38 MAPK pathway. ( |