- Title
-
Haematopoietic stem and progenitor cell heterogeneity is inherited from the embryonic endothelium
- Authors
- Ghersi, J.J., Baldissera, G., Hintzen, J., Luff, S.A., Cheng, S., Xia, I.F., Sturgeon, C.M., Nicoli, S.
- Source
- Full text @ Nat. Cell Biol.
|
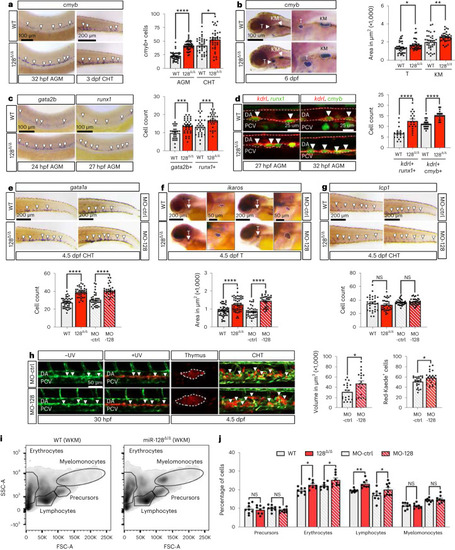
nHSPC development and blood lineages are altered in miR-128Δ/Δ. |
|
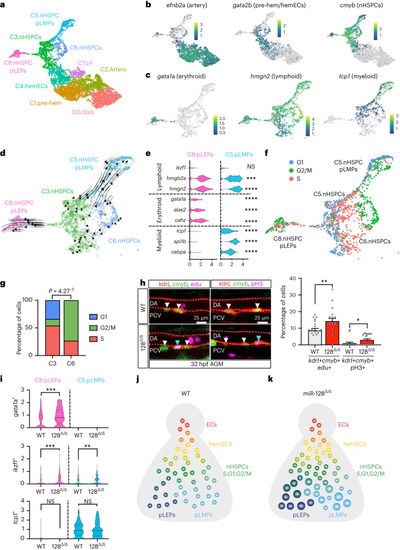
nHSPC heterogeneity is defined by cell cycle and lineage bias phenotypes and regulated by miR-128. |
|
miR-128 function in ECs to regulate HSPC heterogeneity before EHT. |
|
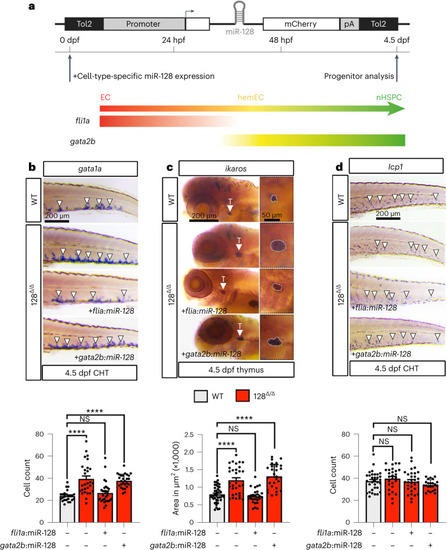
miR-128 function before EHT is conserved in hPSCs. |
|
miR-128 regulation of Notch (via |
|
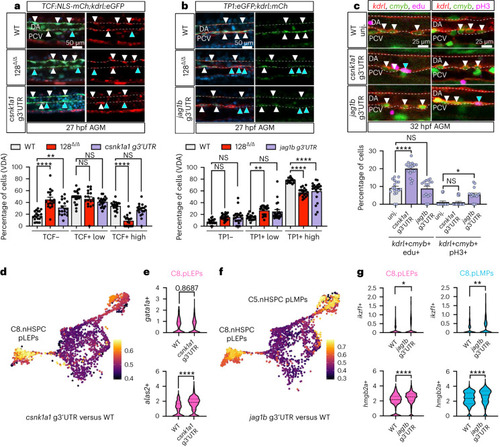
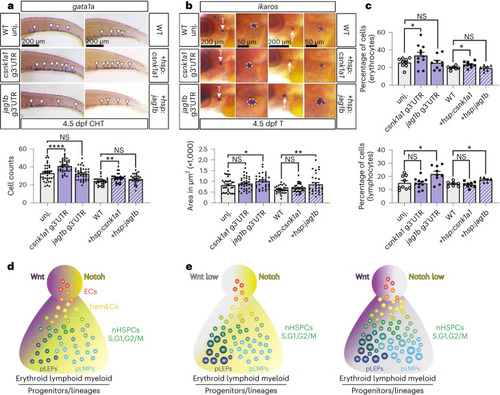
miR-128 regulation of jag1b and csnk1a1 in the embryonic EHT control long-term blood lineages. |