- Title
-
Tissue-Engineered Disease Modeling of Lymphangioleiomyomatosis Exposes a Therapeutic Vulnerability to HDAC Inhibition
- Authors
- Pietrobon, A., Yockell-Lelièvre, J., Melong, N., Smith, L.J., Delaney, S.P., Azzam, N., Xue, C., Merwin, N., Lian, E., Camacho-Magallanes, A., Doré, C., Musso, G., Julian, L.M., Kristof, A.S., Tam, R.Y., Berman, J.N., Shoichet, M.S., Stanford, W.L.
- Source
- Full text @ Adv Sci (Weinh)
|
Hydrogel culture of stem cell‐derived disease models exhibits features of LAM. A) Representative immunofluorescence images of WT and |
|
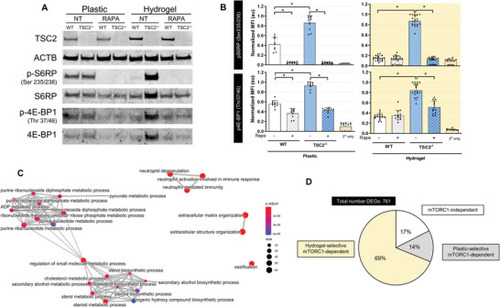
Hydrogel culture potentiates differential mTORC1‐signaling between WT and |
|
3D drug screen identifies HDAC inhibitors as anti‐invasive and selectively cytotoxic toward |
|
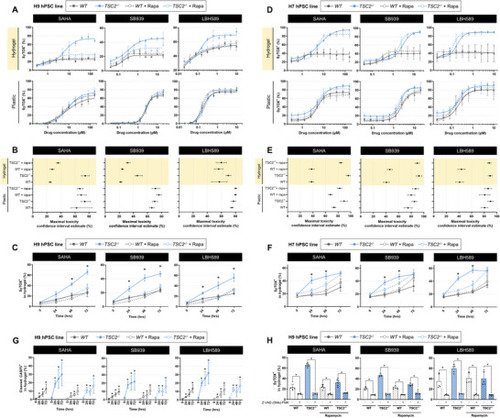
Three safe‐in‐human HDAC inhibitors induce mTORC1‐dependent selective cytotoxicity exclusively in hydrogel culture. A,D) Dose–response cytotoxicity curves of H9 (panel A) and H7 (panel D) cells treated with the indicated HDAC inhibitor for 3 days while cultured on plastic or hydrogel ± 20 × 10−9 |
|
HDAC inhibitors attenuate cell invasion independent of cytotoxicity. A) Live |
|
HDAC inhibitors are anti‐invasive and selectively cytotoxic toward TSC2−/− cells xenotransplanted into zebrafish. A) Representative phase contrast image of 1 day post‐injection (dpi) zebrafish larvae injected with |