- Title
-
Infertility control of transgenic fluorescent zebrafish with targeted mutagenesis of the dnd1 gene by CRISPR/Cas9 genome editing
- Authors
- Chu, W.K., Huang, S.C., Chang, C.F., Wu, J.L., Gong, H.Y.
- Source
- Full text @ Front Genet
|
(A) Design of the CRISPR/Cas9 target sites of the guide RNA (gRNA) in dnd1. The gRNA was designed in exon 2, and the target site of the gRNA is indicated in red, while the sequence highlighted in yellow is PAM. The RNA-recognition motif (RRM) is the conserved functional domain encoded by exon 3 of the dnd1 gene. (B) Microinjection schematic with the component and concentration of the working solution. (C) Mutation identified in each |
|
(A) Mutation sequences identified in the |
|
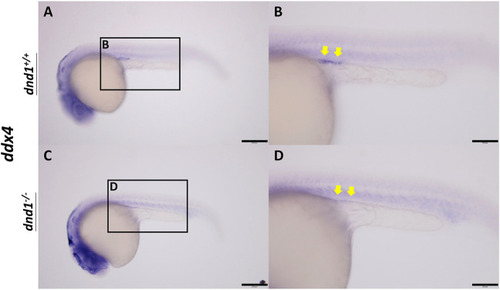
ddx4 expression visualized by whole-mount in situ hybridization in 24-h post-fertilized embryos of the |
|
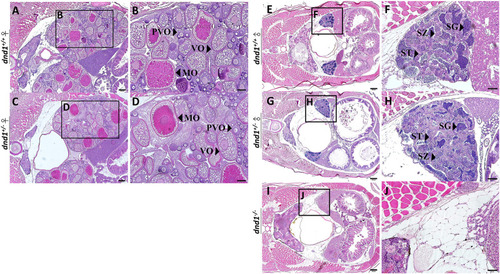
Histological analyses of zebrafish gonad. (A)-(B) Wild-type zebrafish ovary. (C)-(D) Heterozygote zebrafish ovary. (E)-(F) Wild-type zebrafish testis. (G)-(H) Heterozygote zebrafish testis. (I)-(J) Mutant zebrafish testis. PVO: previtellogenic oocytes; VO: vitellogenic oocytes; MO: mature oocyte; SC: spermatocytes; SG: spermatogonia; ST: spermatids. Bars = 200 μm (A, C, E, G, I), 100 μm (B, D) and 50 μm (F, H, J). |
|
Analysis of the expression of germ-cell-related genes between the wild-type ( |
|
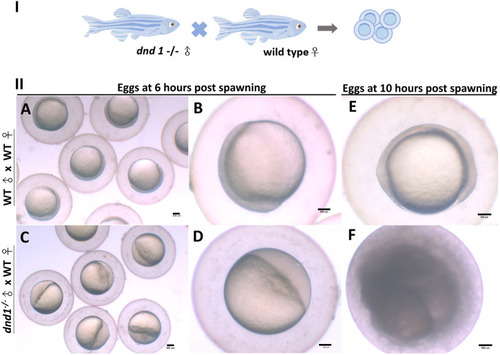
(I) dnd1 −/− zebrafish were all male, with a normal courtship behavior. (II) Spawned eggs from WT female mating with dnd1 −/− male and wild-type zebrafish. (A)-(B) Embryos spawned by wild-type zebrafish after 6 h and (E) after 10 h. (C)-(D) Eggs spawned by mutant male zebrafish and wild-type female zebrafish after 6 h and (F) after 10 h. Bars = 100 μm. |
|
Histological analyses of fluorescent zebrafish testis structure. (A)-(C) dnd1+/+ fluorescent zebrafish. (D)-(F) dnd1-knockout homozygote fluorescent zebrafish. Bars = 500 μm (A, D), 200 μm (B, E), and 20 μm (C, F). |
|
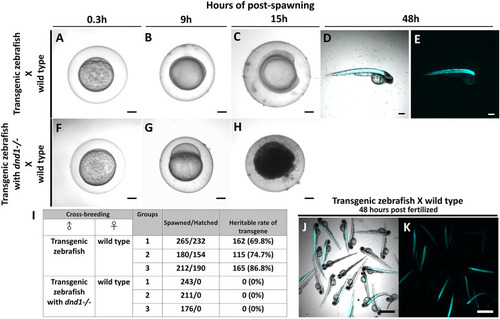
Transgenic zebrafish with dnd1 mutant can effectively avoid the transgene inherited. The eggs spawned by transgenic zebrafish and wild type zebrafish (A) .3 h (B) 9 h (C) 15 h (D) 48 h of post spawning under light view (E) 48 h of post spawning under dark view. The eggs spawned by transgenic zebrafish with dnd1 mutant and wild type zebrafish (F) .3 h (G) 9 h (H) 15 h. (I) The heritable rate counting of transgene in cross-breeding group. Top view of hatched offspring of cross-breeding by transgenic zebrafish and wild type zebrafish. (J) light view (K) dark view. Bars = 200 μm (A–H) and 500 μm (J, K). |
|
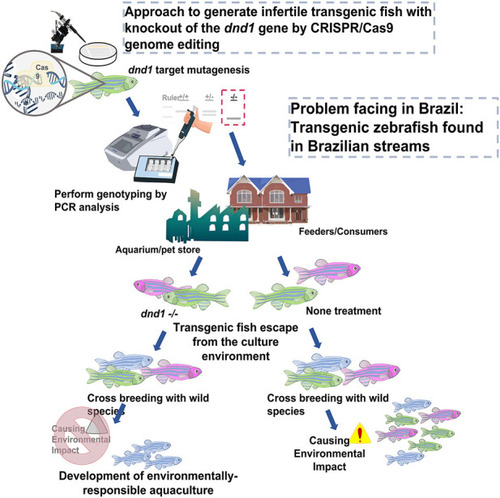
Approach to generate infertile transgenic fluorescent zebrafish with knockout out of the dnd1 gene by CRISPR/Cas9 genome editing. We demonstrate that the dnd1 knockout by CRISPR/Cas9 genome editing could be applied to the infertility control of transgenic fluorescent zebrafish to prevent them from cross-breeding with wild species when they escape into the wild field. It can promote the development of an environmentally responsible aquaculture. |