- Title
-
A novel uveitis model induced by lipopolysaccharide in zebrafish
- Authors
- Xiao, X., Liu, Z., Su, G., Liu, H., Yin, W., Guan, Y., Jing, S., Du, L., Li, F., Li, N., Yang, P.
- Source
- Full text @ Front Immunol
|
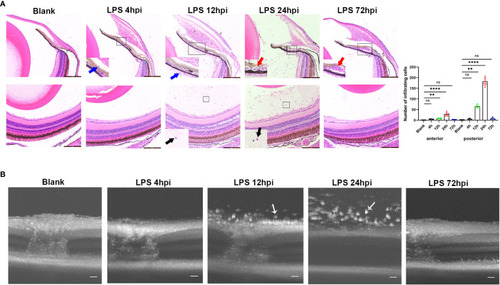
Inflammatory sign of EIU in zebrafish over time-course. |
|
Distribution of immune cells during EIU inflammation process in zebrafish. |
|
Transcriptomic traits of EIU in zebrafish. |
|
RT-PCR validation of DEGs in the initial of inflammation (mean ± SD; * |
|
Prednisone immersion treatment inhibited EIU inflammation in zebrafish. |