- Title
-
Comprehensive Mapping and Dynamics of Site-Specific Prolyl-Hydroxylation, Lysyl-Hydroxylation and Lysyl O-Glycosylation of Collagens Deposited in ECM During Zebrafish Heart Regeneration
- Authors
- Sarohi, V., Srivastava, S., Basak, T.
- Source
- Full text @ Front Mol Biosci
|
Identification of collagen chains and their relative abundances in zebrafish heart ECM: (A) Total number of collagen chains (Cabral et al., 2007) identified previously by Garcia-Puig et al. compared to (Padmanabhan Iyer et al., 2016) number of collagen chains identified in our analysis. (B) depicts inclusion of hydroxyproline (HyP) modification in the database search by MyriMatch and MSFragger resulting in identification of almost 61.12 and 40.46% (summed number from all the raw *.pepXML files used for database search) new unique peptides. This strategy yielded more no. of peptide identification resulting in a higher number of collagen chain identification from the same dataset. (C) Top 10 abundant collagen chains deposited in the zebrafish heart ECM were identified by two different search engines MyriMatch and MSFragger respectively. (D) Relative abundances of different collagen chains during zebrafish heart regeneration are shown by the heatmap. Light yellow represents the lower value (−2) and dark red represents the higher value (+2) in the row. Normalized spectral count values have been used to generate the heatmap (considering ≥ 3 spectral counts per chain). collagen chains marked with red (.) dots are quantitated during regeneration in re-analysis for the first time (DPA = day post amputation). |
|
Optimized dual database search engine-based MS analysis pipeline for the global identification and quantitation of site-specific collagen PTMs from zebrafish heart ECM. Thermo. raw or Bruker. d MS/MS files were initially converted to. mgf and. mzML files (by MSConvert) respectively and searched with MyriMatch and MSFragger to identify the collagen present in the zebrafish heart ECM. For MyriMatch, the subset of identified proteins was used as a second database to perform a PTM module enabled search defining specific sequence motifs for the site-specific identification of collagen PTMs in zebrafish heart ECM. For MSFragger, the PTM searches were conducted directly with the entire zebrafish database. From MyriMatch and MSFragger *.pep.XML output files containing each peptide spectrum match (PSM) were further parsed by PeptideProphet to compute the probability score (0,1). The *.pep.XML output file parsed by PeptideProphet was further imported into Skyline along with all the raw MS/MS files in to generate the spectral library (.blib). This spectral library (.blib) in Skyline was used for the targeted MS1-based extraction of all the PTM modified and unmodified collagen peptide species for each specific site. The area of MS1 area for each peptide for different samples was computed from Skyline. |
|
Comprehensive map of proline/lysine hydroxylation sites and lysine O-glycosylation sites in COL1A1a of WT zebrafish heart ECM. Identified peptide sequence in the proteomic analysis is shown in black color, sequence not identified in this analysis are shown in grey color. A total of 94.98% sequence coverage of COL1A1a is detected (considering the matured form of COL1A1The signalgnal peptide is 22 amino acids (1–22) long. Sequence alignment matching with human COL1A1 revealed the propeptide cleavage sites. Dark yellow arrows show N terminal (23–146) and C-Term (1,202–1,447) propeptide cleavage sites. As shown in the top right corner, red bold “P” with a blue star represents 3-hydroxyproline on the Xaa position followed by 4-hydroxyproline the on Yaa position in the Gly-Xaa-Yaa motif. 4-hydroxyproline on Yaa position is represented with red color “P”. Hydroxyproline on unusual Xaa position with (Ala, Val, Met, Ile, Ser, Glu, Arg, and Asp) on Yaa position are also identified but cannot label either 3-hydroxyproline or 4-hydroxyproline. Hydroxylysine sites are presented by bold “K”. Lysine sites highlighted with a yellow circle represents galactosyl-hydroxylysine sites and yellow plus blue coloued circles represent glucosylgalactosyl-hydroxylysine sites. The presence of glucosylgalactosyl-hydroxylysine, galactosyl-hydroxylysine, and hydroxylysine on the same site shows lysine microheterogeneity. A summary of these site-specific PTMs of COL1A1a is presented in Table 1, and all the PSMs for O-glycosylated lysine and 3-hydroxyproline sites are provided in Supplementary Figure S2.1-S2.21. |
|
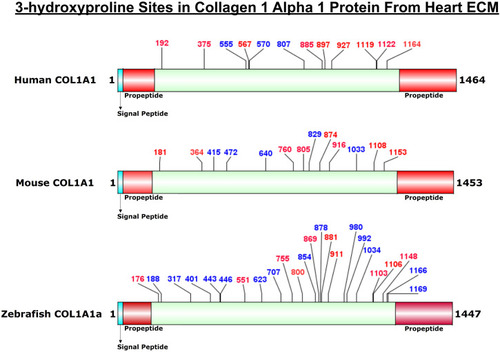
Comparison of 3-HyP sites identified in COL1A1a from zebrafish heart ECM to COL1A1 of human and mice heart ECM. The horizontal box represents the full-length COL1A1 sequence and the vertical black lines indicate the corresponding 3-HyP sites. The information for 3-HyP sites of human and mice heart ECM were re-analyzed in this manuscript from the available raw MS data from Barallobre-Barreiro et al. and Padmanabhan et al. The 3-HyP sites marked with red represent the conserved sites among human, mouse, and zebrafish. |
|
Comprehensive PTM map of COL1A1b of WT zebrafish heart ECM, presenting proline/lysine hydroxylation sites and lysine O-glycosylation sites. The representation of PTM sthe ites is similar to the COL1A1a PTM map as shown in top right corner. Peptides identified in proteomics analysis are shown in black color and unidentified peptides are shown in grey color. A total of 96.48% sequence coverage of COL1A1a is detected (considering the matured form of COL1A1b). The signal peptide is 22 amino acids (1–22) long. Sequence alignment matching and previous analysis by Gistelink et al. with human COL1A1 revealed the propeptide cleavage sites. Dark yellow arrows show N terminal (23–150) and C-Term (1,204–1,447) propeptide cleavage sites. Red bold “P” with a blue star represents 3-HyP and red “P” represents 4-HyP. Hydroxylysine is represented with bold “K” and yellow and blue circle represents the lysine O-glycosylation. A summary of these site-specific PTMs of COL1A1b is presented in Table 1, and all the PSMs for O-glycosylated lysine and 3-hydroxyproline sites are provided in Supplementary Figures. S2.22–S2.52. |
|
Comprehensive map of COL1A2 of ECM of WT zebrafish heart. It presents proline/lysine hydroxylation sites and lysine glycosylation sites. Representation of PTM sites is similar to COL1A1a and COL1A1b PTM maps. Peptides identified in proteomics analysis are shown in black color and unidentified peptides are shown in grey color. Total 97.21% sequence coverage of COL1A2 is detected (considering the matured form of COL1A1b). The signal peptide is 22 amino acids (1–22) long. N terminal propeptide (23–68) and C terminal propeptide (1,109–1,352) cleavage sites are marked with dark yellow arrows. Red bold “P” with blue star represents 3-HyP and red “P” represents 4-HyP. Hydroxylysine is represented with bold “K” and yellow and blue circle represents the lysine O-glycosylation. A summary of these site-specific PTMs of COL1A2 is presented in Table 1, and all the PSMs for O-glycosylated lysine and 3-hydroxyproline sites are provided in Supplementary Figure S2.53–S2.83. |
|
Heatmap depicting the relative occupancy level of 3-hydroxyproline sites in three different chains of collagen 1 deposited in zebrafish heart ECM during regeneration. Further occupancy of one 3-HyP cluster of COL5A2a1195, 1201 was also quantitated during zebrafish heart regeneration. Normalized occupancy values of prolyl-3-hydroxylations were computed to generate the heat map. Light yellow represents the low value (lowest occupancy) and dark red shows the higher value (highest occupancy) in the row. |
|
Quantitation of microheterogeneity of K1017 site in COL1A1a present in zebrafish heart ECM during regeneration. (A) Chromatogram plots represent the elution of unmodified (K), hydroxylysine (HyK), and glucosylgalactosyl-hydroxylysine (GG-HyK) modified K1017 site containing peptide 1011DGAAGPKGDRGETGPSGTPGAPGPPGAAGPIGPAGK1046 (B) Graphical representation of Skyline-based MS1 quantitation of micro-heterogenic distribution of unmodified K1017 (yellow), HyK1017 (green) and GG-HyK277 (blue) species in COL1A1a from the ECM digest of control, 7 DPA, 14 DPA, and 30 DPA regenerating zebrafish heart. The different colors in the bar represent the occupancy of different forms at the K1017 site in COL1A1a with mean ± SEM. An increase in glucosylgalactosyl-hydroxylysine levels during regeneration is significant (ANOVA, p < 0.05) (See Table 3) |