- Title
-
Intranasal delivery of SARS-CoV-2 Spike protein is sufficient to cause olfactory damage, inflammation and olfactory dysfunction in zebrafish
- Authors
- Kraus, A., Huertas, M., Ellis, L., Boudinot, P., Levraud, J.P., Salinas, I.
- Source
- Full text @ Brain Behav. Immun.
|
|
|
|
|
|
|
|
|
|
|
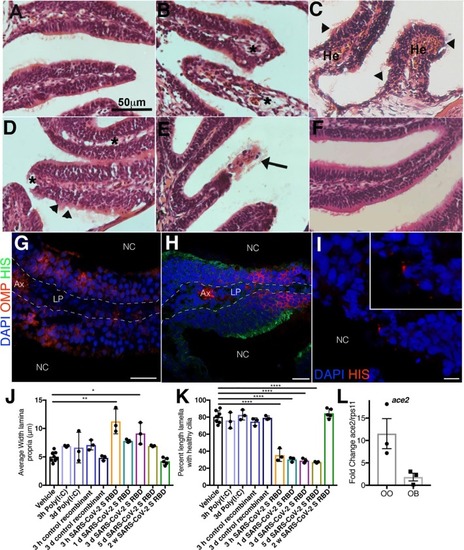
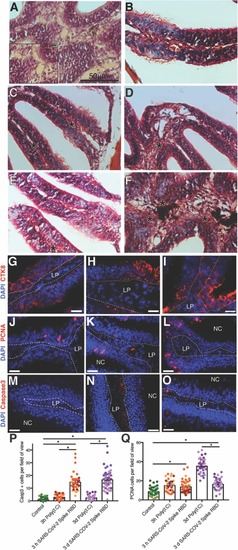
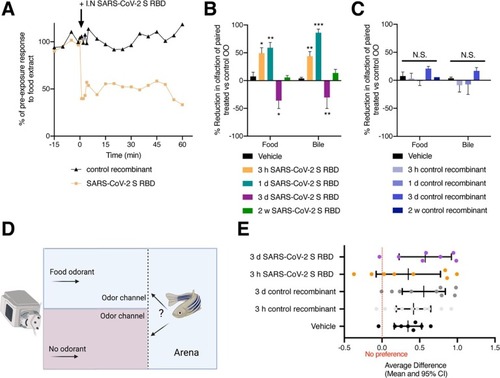
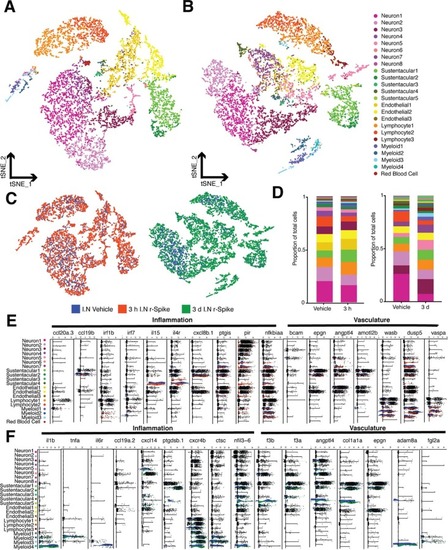
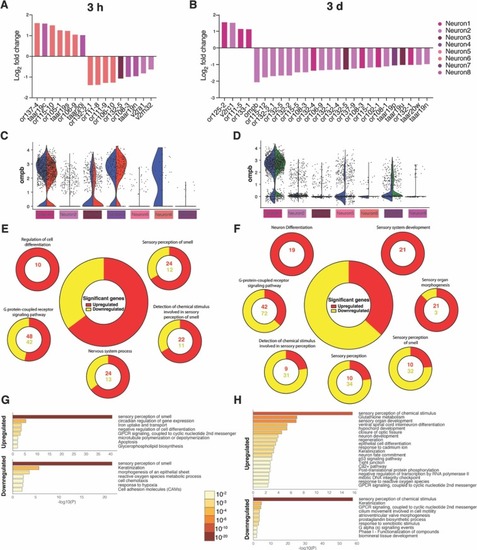
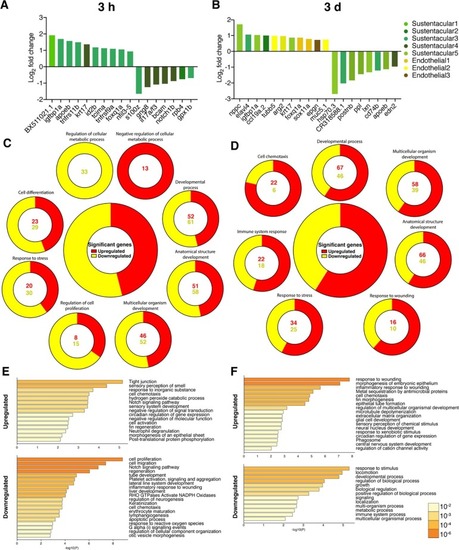
Fig. 6. Intranasal SARS-CoV-2 S RBD protein delivery causes transcriptional changes in zebrafish sustentacular and endothelial clusters. (A-B) Bar plots showing fold-change in expression of selected significantly differentially expressed genes in sustentacular and endothelial cell clusters in the zebrafish OO 3 h and 3 d post intranasal SARS-CoV-2 S RBD protein compared to vehicle controls identified by scRNA-Seq. (C-D) Summary webs of gene ontology analysis performed in ShinyGO v0.61 from the scRNA-Seq data showing the number of genes significantly up and down regulated in sustentacular and endothelial cell clusters in the OO of zebrafish 3 h and 3 days post-intranasal SARS-CoV-2 S RBD protein compared to vehicle treated controls. (E-F) Gene set enrichment analysis using Metascape of genes from sustentacular and endothelial clusters that were either significantly up or down regulated in zebrafish OO 3 h and 3 d post-intranasal SARS-CoV-2 S RBD protein compared to vehicle treated controls (adjusted P-value < 0.05). Significantly enriched terms are colored by P-value. |