- Title
-
WhiB4 Is Required for the Reactivation of Persistent Infection of Mycobacterium marinum in Zebrafish
- Authors
- Lin, C., Tang, Y., Wang, Y., Zhang, J., Li, Y., Xu, S., Xia, B., Zhai, Q., Li, Y., Zhang, L., Liu, J.
- Source
- Full text @ Microbiol Spectr
|
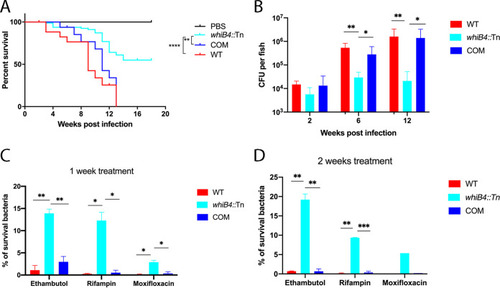
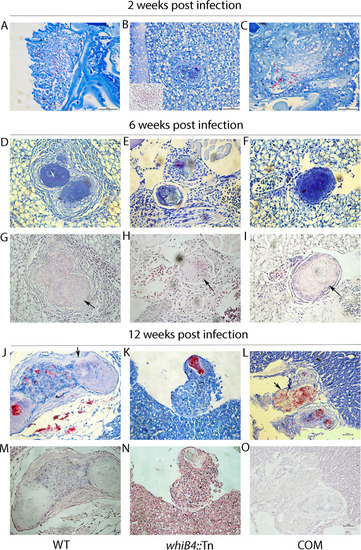
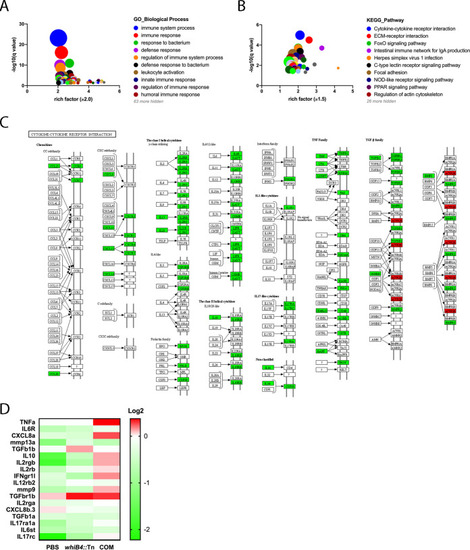
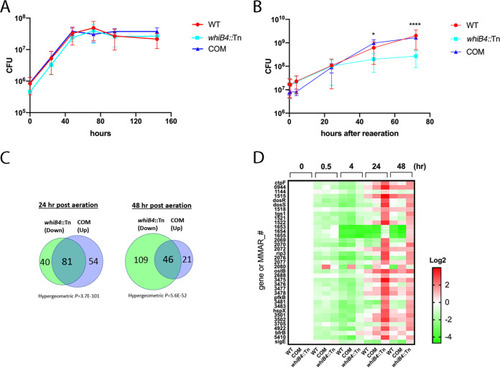
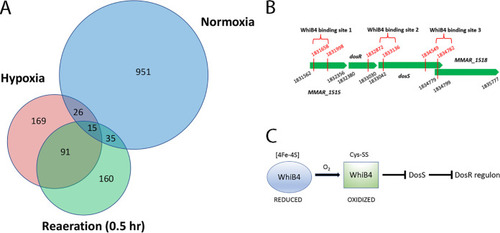
(A and B) The w PHENOTYPE:
|
|
The |
|
The |
|
The |
|
WhiB4 binds |