- Title
-
Chondroitin/dermatan sulfate glycosyltransferase genes are essential for craniofacial development
- Authors
- Habicher, J., Varshney, G.K., Waldmann, L., Snitting, D., Allalou, A., Zhang, H., Ghanem, A., Öhman Mägi, C., Dierker, T., Kjellén, L., Burgess, S.M., Ledin, J.
- Source
- Full text @ PLoS Genet.
|
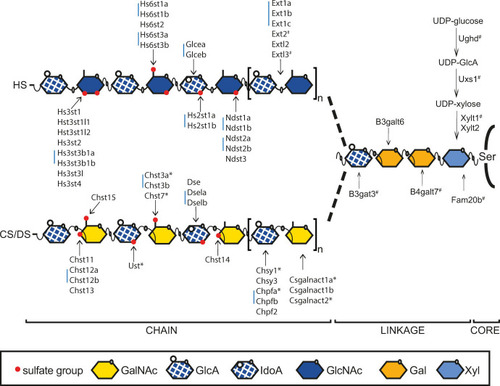
Heparan sulfate (HS) and chondroitin/dermatan sulfate (CS/DS) biosynthesis in zebrafish.
HS and CS/DS chains are attached to serine (Ser) residues of the core protein. The first four monosaccharides form the linkage region of both HS and CS/DS. Extl3 initiates HS polymerization, while Csgalnact1a and Csgalnact2 perform this function in CS/DS polymerization. Elongation of HS is carried out by Ext enzymes, while Chsy and Chpf enzymes polymerize the CS/DS chain. The first modification of HS is carried out by Ndst enzymes, replacing an N-acetyl groups of GlcNAc residues with an N-sulfate group. Glcea and Glceb epimerize GlcA into IdoA in HS, while Dse, Dsela and Dselb are responsible for this modification in DS. Hs2st enzymes add a sulfate groups to the IdoA C-2 position of HS and Hs6st and Hs3st enzymes add sulfate groups to the GlcNAc or GlcNS residues. Ust adds a sulfate group to the hexuronic acid C-2 position of CS/DS while Chst11, Chst12a, Chst12b, Chst13 and Chst14 are GalNAc 4-O-sulfotransferases while Chst3a, Chst3b, Chst7 and Chst15 are GalNAc 6-O-sulfostransferases. Blue bars indicate two or three zebrafish genes orthologous to a single mammalian gene. Ndst3 in zebrafish is a single gene orthologous to two mammalian genes (NDST3 and NDST4). # Indicates previously published mutant zebrafish alleles while * indicates the targeted mutations reported in this study. Xyl: xylose, Gal: galactose, GlcNAc: N-acetylglucosamine, IdoA: iduronic acid, GlcA: glucuronic acid, GalNAc: N-acetylgalactosamine. |
|
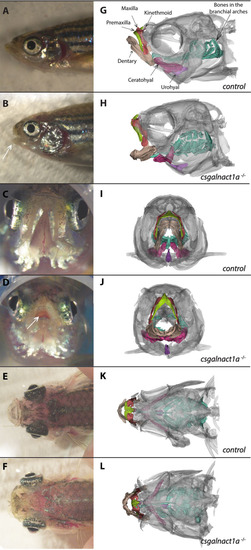
Csgalnact1a-/- adults display obvious craniofacial malformations.
Bright field images of zebrafish heads (A-F) and corresponding 3D reconstructions of microCT scans (G-H). Different skeletal elements are highlighted with colors. Compared to control adults (A,C,E,G,I,K), csgalnact1a-/- adults (B,D,F,H,J,L) present a shortened jaw (arrow B) resulting in an open mouth phenotype (arrow D). In dorsal view the protruding eyes of csgalnact1a-/- adults are apparent (F). |
|
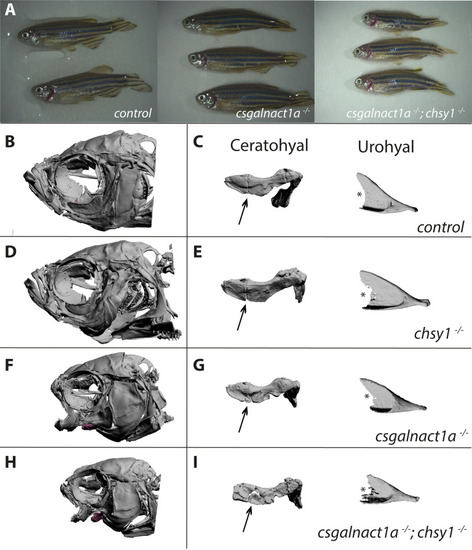
Severe malformation in the adult head skeleton.
Alterations in the morphology of the ceratohyal and urohyal were magnified in all four genotypes (for spatial position of elements in the zebrafish head, see S1 movie). Adult csgalnact1a-/- ; chsy1-/- zebrafish displayed gross malformations and reduced body size compared to control and csgalnact1a-/- adults (A). 3D reconstructions of all genotypes showed that no bone structures were missing (B,D,F,H and S1–S4 Figs). However, shorter and misshaped elements were common in csgalnact1a-/- (F) and csgalnact1a-/- ; chsy1-/- fish (H), while the chsy1-/- skeletal phenotype (D) was milder, compared to the control (B). The differences in morphological integrity of skeletal elements was evident from magnifications, for example the boundary between the anterior and the posterior part (arrow) of the ceratohyal and posterior edge (*) and of the urohyal (C,E,G,I and S5-12). |
|
Malformations in the craniofacial cartilage structures.
Maximum projections of average patterns generated from control, csgalnact1a-/- and chsy1-/- alcian blue stained larvae at 9 dpf show the pharyngeal cartilage structures in a ventral view (A-F). Maximum projections for each mutant (magenta) aligned to the control (green) are displayed using a rigid transformation (G-J). Measurements of the length and width of the head skeleton of 9 dpf larvae were performed on maximum projection images as shown in the image to the right (K) and plotted as a factor of control larvae (n = 8 for all genotype groups) (K). csgalnact1a-/- and chsy1-/- larval head skeleton is significantly smaller compared to control (K). Measurements of the standard body length (1) and different other measurements of the head (2–5) are indicated on images of 40 dpf old juvenile fish (L). chsy1-/- juveniles are significantly shorter and have a smaller head compared to control larvae (chsy1-/- n = 17, control n = 27) (L). Statistical significance is indicated by * for p-values <0.05 and ** for p-values <0.005. |
|
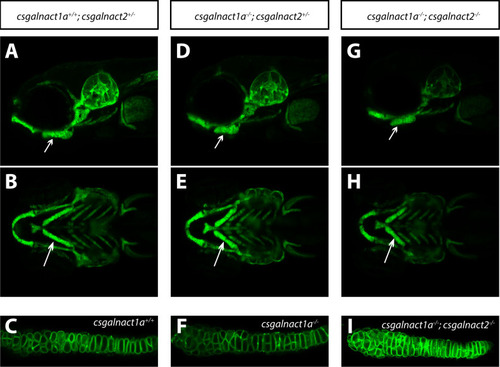
Craniofacial malformations in csgalnact1a-/- and csgalnact1a-/-; csgalnact2-/- larvae.
LightSheet images of live Tg(col2a1a:mEGFP) larvae at 6 dpf show craniofacial cartilage elements formed in csgalnact1a+/+;csgalnact2+/- (A,B,C), csgalnact1a-/-;csgalnact2+/- (D,E,F) and csgalnact1a-/-;csgalnact2-/- larvae (G,H,I). Lateral views (A,D,G) display a shorter ceratohyal, ventral views display a wider angle between the ceratohyal elements in csgalnact1a-/-;csgalnact2+/- (E) and csgalnact1a-/-;csgalnact2-/- (G) larvae (arrow) compared to controls (A,B). A detailed view on the chondorcytes within the ceratohyal (arrow) shows that intercalation occurs even in csgalnact1a-/- (F) and csgalnact1a-/-;csgalnact2-/- (I) larvae. |
|
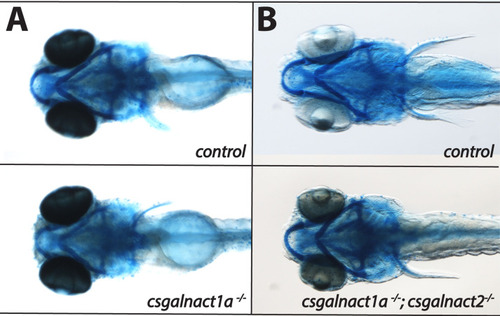
Larvae at 6 dpf were stained with alcian blue and are displayed in ventral views.
No significant reduction of alcian blue staining was detected in either in the csgalnact1a-/- (A) nor in the csgalnact1a-/-;csgalnact2-/- (B) larvae. |
|
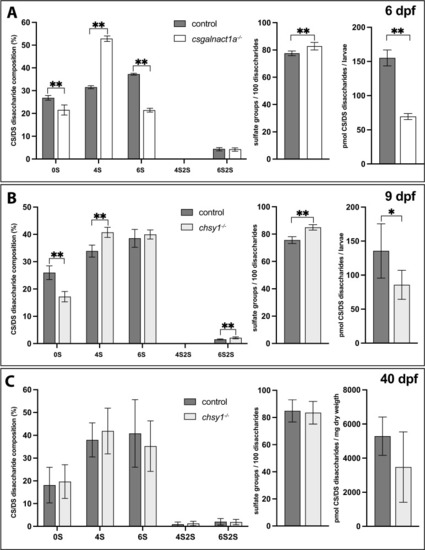
RPIP-HPLC analysis shows the CS/DS disaccharide composition (left charts), total sulfate group content/100 disaccharides (middle charts) and amount of CS/DS (right charts) for 6 dpf larvae (A), 9 dpf larvae (B) and 40 dpf juveniles (C). Statistical significances are indicated with * for p-values <0.05 and ** for p-values <0.005. Disaccharide species abbreviated as 0S (ΔHexA- GalNAc/ΔHexA-GlcNAc), 4S (ΔHexA-GalNAc4S), 6S (ΔHexA-GalNAc6S), 4S2S (ΔHexA2S-GalNAc4S), and 6S2S (ΔHexA2S-GalNAc6S). |
|
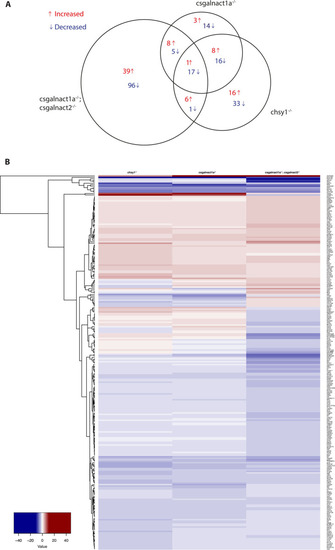
The area proportional venn diagram (A) shows differentially expressed genes in csgalnact1a-/-*, csgalnact1a-/-;csgalnact2-/- and chsy1-/- larvae compared to control larvae. The heat map (B) shows all genes with a two-fold increase (red) or decrease (blue) in the csgalnact1a-/-, csgalnact1a-/-;csgalnact2-/- or chsy1-/- larvae. * 50% of the csgalnact1a-/- individuals lack one functional allele of chsy1. |