- Title
-
Interspecies Differences in Activation of Peroxisome Proliferator-Activated Receptor γ by Pharmaceutical and Environmental Chemicals
- Authors
- Garoche, C., Boulahtouf, A., Grimaldi, M., Chiavarina, B., Toporova, L., den Broeder, M.J., Legler, J., Bourguet, W., Balaguer, P.
- Source
- Full text @ Env. Sci. Tech.
|
Figure 1. Chemical structures of the molecules used in this study. * NRs antagonists. |
|
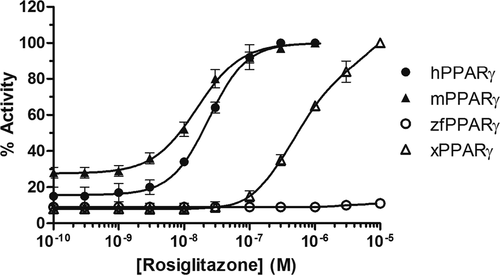
Figure 2. Transcriptional activity of h, m, zf, and xPPARγ in response to rosiglitazone. Results are expressed as a percentage of the maximum luciferase activity induced by 1 μM rosiglitazone (HG5LN-hPPARγ, HG5LN-mPPARγ), 10 μM GW 3965 (HG5LN-zfPPARγ), or 10 μM rosiglitazone (HG5LN-xPPARγ). Error bars represent standard deviations. |
|
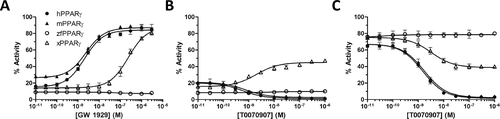
Figure 3. Transcriptional activity of h, m, zf, and xPPARγ in response to synthetic hPPARγ ligands. Results are expressed as a percentage of the maximum luciferase activity induced by 1 μM rosiglitazone (HG5LN-hPPARγ, HG5LN-mPPARγ), 10 μM GW 3965 (HG5LN-zfPPARγ), or 10 μM rosiglitazone (HG5LN-xPPARγ). GW 1929 (A) was tested in agonist assays; T0070907 was tested in agonist (B) and antagonist (C) assays. Error bars represent standard deviations. |
|
Figure 4. Transcriptional activity of h, m, zf, and xPPARγ in response to synthetic hPPARα and hLXR ligands. Results are expressed as a percentage of the maximum luciferase activity induced by 1 μM rosiglitazone (HG5LN-hPPARγ, HG5LN-mPPARγ), 10 μM GW 3965 (HG5LN-zfPPARγ), or 10 μM rosiglitazone (HG5LN-xPPARγ). Agonist hPPARα ligand GW 7647 (A) tested in agonist assay; antagonist hPPARα ligand GW 6471 (B) tested in antagonist assay; and agonist hLXR ligand GW 3965 tested in agonist assays (C). Error bars represent standard deviations. |
|
Figure 5. Transcriptional activity of h, m, zf, and xPPARγ in response to environmental compounds. Results are expressed as a percentage of the maximum luciferase activity induced by 1 μM rosiglitazone (HG5LN-hPPARγ, HG5LN-mPPARγ), 3 μM GW 3965 (HG5LN-zfPPARγ), or 10 μM rosiglitazone (HG5LN-xPPARγ). Compounds were tested without serum. Error bars represent standard deviations. |
|
Figure 6. Structural basis for differential ligand-binding specificities of h, x, and zfPPARγ. (A) Rosiglitazone is positioned in the ligand-binding pocket of hPPARγ (crystal structure PDB code 2PRG). Residues that differ in the ligand-binding pockets of hPPARγ (orange) and zfPPARγ (blue) are displayed and labeled. The zebrafish receptor was modeled using the server EDMon (http://edmon.cbs.cnrs.fr). hPPARγ G312 and C313, which are replaced by serine and tyrosine residues, respectively, in the zebrafish receptor, are highlighted with red labels. (B) Superposition of the crystal structure of hPPARγ (2PRG) bound to rosiglitazone and a model of xPPARγ generated by the server EDMon. Residues that differ in the ligand-binding pockets of hPPARγ (orange) and xPPARγ (magenta) are displayed and labeled (except the tyrosine residue in helix H12, which is conserved in both species). hPPARγ C313 and S317, which are replaced by leucine residues in the xenopus receptor, are highlighted with red labels. (C) TPP as modeled in the ligand-binding pocket of hPPARγ. |