- Title
-
Hundreds of LncRNAs Display Circadian Rhythmicity in Zebrafish Larvae
- Authors
- Mishra, S.K., Zhong, Z., Wang, H.
- Source
- Full text @ Cells
|
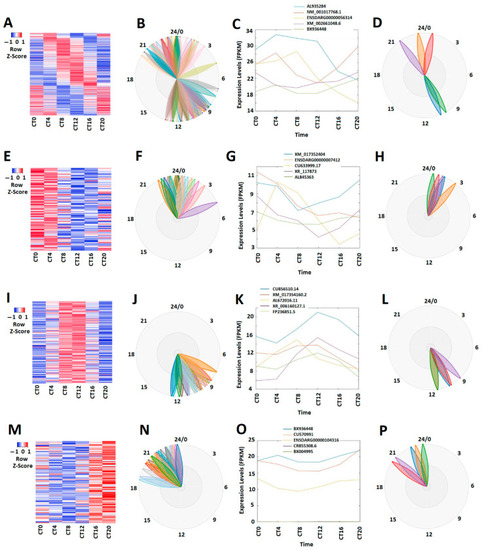
Expression profile analysis of circadianly expressed zebrafish larval lncRNAs under the DD condition. (A–D) Analysis of all the 269 circadianly expressed larval lncRNAs under the DD condition. Heat map (A) and BioDare2 plots (B) of all the 269 circadianly expressed zebrafish larval lncRNAs, expression profiles (C) and BioDare2 plots (D) of representative lncRNAs. (E–H) Analysis of 100 larval morning (CT0 and CT4) lncRNAs. Heat map (E) and BioDare2 plots (F) of the 100 larval zebrafish morning lncRNAs, expression profiles (G) and BioDare2 plots (H) of zebrafish larval morning representative lncRNAs. (I–L) Analysis of 75 zebrafish larval evening (CT8 and CT12) lncRNAs. Heat map (I) and BioDare2 plots (J) of the 75 larval evening lncRNAs, expression profiles (K) and BioDare2 plots (L) of larval evening representative lncRNAs. (M–P) Analysis of 94 larval zebrafish night (CT16 and CT20) lncRNAs. Heat map (M) and BioDare2 plots (N) of the 94 larval zebrafish night lncRNAs, expression profiles (O) and BioDare2 plots (P) of larval zebrafish night representative lncRNAs. |
|
GO, COG, and KEGG analyses of circadianly expressed larval zebrafish lncRNAs under the DD condition. (A) GO annotation of the zebrafish larval lncRNAs under the DD condition revealed their potential involvement in several biological processes, such as nuclear receptor transcription coactivator activity, regulation of response to DNA damage stimulus, and regulation of DNA metabolic process. (B) COG functional classification of circadianly expressed larval zebrafish lncRNAs under the DD condition. (C) KEGG pathway enrichment analysis of the circadianly expressed larval zebrafish lncRNAs under the DD condition. |
|
Expression profile analysis circadianly expressed zebrafish larval lncRNAs under the LL condition. (A–D) Analysis of all the 309 circadianly expressed larval lncRNAs under the DD condition. Heat map (A) and BioDare2 plots (B) of all 309 the circadianly expressed larval lncRNAs, expression profiles (C) and BioDare2 plots (D) of representative lncRNAs. (E-H) Analysis of the 78 larval morning (CT0 and CT4) lncRNAs. Heat map (E) and BioDare2 plots (F) of the 78 larval morning lncRNAs, expression profiles (G) and BioDare2 plots (H) of larval morning representative lncRNAs. (I–L) Analysis of 144 larval evening (CT8 and CT12) lncRNAs. Heat map (I) and BioDare2 plots (J) of 144 larval evening lncRNAs, expression profiles (K) and BioDare2 plots (L) of larval evening representative lncRNAs. (M–P) Analysis of 87 larval night (CT16 and CT20) lncRNAs. Heat map (M) and BioDare2 plots (N) of 87 larval night lncRNAs, expression profiles (O) and BioDare2 plots (P) of larval night representative lncRNAs. |
|
GO, COG, and KEGG analyses of circadianly expressed zebrafish larval lncRNAs under the DD condition. (A) GO annotation revealed the lncRNAs potentially involved in different biological processes, such as nuclear-transcribed mRNA catabolic process, and eye photoreceptor cell differentiation. (B) COG functional classification of circadianly expressed larval lncRNAs under the DD condition. (C), KEGG pathway enrichment analysis of circadianly expressed lncRNAs under the DD condition. |
|
Expression profile analysis of circadianly expressed zebrafish larval lncRNAs coexpressed under both the DD condition and the LL condition. (A–D) Analysis of all the 30 coexpressing larval lncRNAs with expression profiles from DD condition. Heat map (A) and BioDare2 plots (B) of all the 30 coexpressing larval lncRNAs, expression profiles (C) and BioDare2 plots (D) of representative lncRNAs. (E–H) Analysis of the 11 coexpressing larval morning (CT0 and CT4) lncRNAs. Heat map (E) and BioDare2 plots (F) of the 11 larval morning lncRNAs, expression profiles (G) and BioDare2 plots (H) of larval morning representative lncRNAs. (I–L) Analysis of the eight coexpressing larval evening (CT8 and CT12) lncRNAs. Heat map (I) and BioDare2 plots (J) of eight larval evening lncRNAs, expression profiles (K) and BioDare2 plots (L) of larval evening representative lncRNAs. (M–P) Analysis of eight coexpressing larval night (CT16 and CT20) lncRNAs. Heat map (M) and BioDare2 plots (N) of eight larval night lncRNAs, expression profiles (O) and BioDare2 plots (P) of larval night representative lncRNAs. |
|
GO, COG, and KEGG analyses of circadianly expressed zebrafish larval lncRNAs coexpressed under both DD and LL conditions. (A) GO annotation revealed the lncRNAs potentially involved in different biological processes, such as cellular protein modification process, protein modification process, cellular macromolecule metabolic process, and protein metabolic process. (B) COG functional classification of circadianly expressed zebrafish larval lncRNAs coexpressed under both DD and LL conditions. (C) KEGG pathway enrichment analysis of circadianly expressed zebrafish larval lncRNAs coexpressed under both DD and LL condition. |
|
Circadianly expressed zebrafish larval lncRNAs are coexpressed in the zebrafish pineal gland and testis. (A) Circadianly expressed lncRNAs coexpressed in larvae, pineal gland and testis under DD condition. (B) Circadianly expressed lncRNAs coexpressed in larvae, pineal gland and testis under LL condition. (C–E) Expression profiles of representative lncRNAs under DD condition: three zebrafish larval morning (CT0 and CT 4) lncRNAs (C), two zebrafish larval evening (CT8 and CT12) lncRNAs (D), and four zebrafish larval night (CT16 and CT20) lncRNAs (E). (F–H) Expression profiles of representative lncRNAs under LL condition: five zebrafish larval morning lncRNAs (F), four zebrafish larval evening lncRNAs (G), and three zebrafish larval night lncRNAs (H). |
|
Three-dimensional structures predicted by the (PS)2-v2 Protein Structure Prediction Server for the lncRNA-encoded peptides simultaneously observed in zebrafish larvae, pineal gland and testis. Zebrafish lncRNA-encoded peptides coexpressed in larvae (DD condition), pineal gland, and testis (A–C), and Zebrafish lncRNA-encoded peptide coexpressed in larvae (LL condition), pineal gland, and testis (D). The 3D structures of the peptides depict the presence of α-helix (pink or purple motif structure), β-strand (yellow layered band), and random coils (white or blue thread) among the coexpressing lncRNA- encoded peptides with the known domains from Protein Data Bank such as, A (1vg5A), B (1gxrA), C (2iftA), and D (1kpfA). |
|
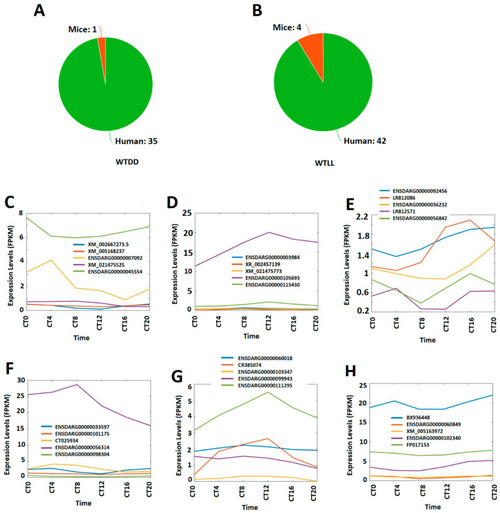
Conservation of circadianly expressed larval lncRNAs in humans and mice, and classification of lncRNAs conserved with humans into the morning, evening, and night groups. The numbers of human and mouse orthologs of the circadianly expressed zebrafish larval lncRNAs under DD condition (A) and LL condition (B). (C–E) Expression profiles of the representative larval morning (CT0 and CT4) lncRNAs (C), evening (CT8 and CT12) lncRNAs (D), and night (CT16 and CT20) lncRNAs (E), under the DD condition, shared with humans. (F–H) Expression profiles of the representative larval morning (CT0 and CT4) lncRNAs (F), evening (CT 8 and CT12) lncRNAs (G), and night (CT16 and CT20) lncRNAs (H), under the LL condition, shared with humans. |