- Title
-
Inhibitory Role of an Aeromonas hydrophila TIR Domain Effector in Antibacterial Immunity by Targeting TLR Signaling Complexes in Zebrafish
- Authors
- Tang, H.P., Huang, C., Hu, C.B., Li, H., Shao, T., Ji, J.F., Bai, J., Fan, D.D., Lin, A.F., Xiang, L.X., Shao, J.Z.
- Source
- Full text @ Front Microbiol
|
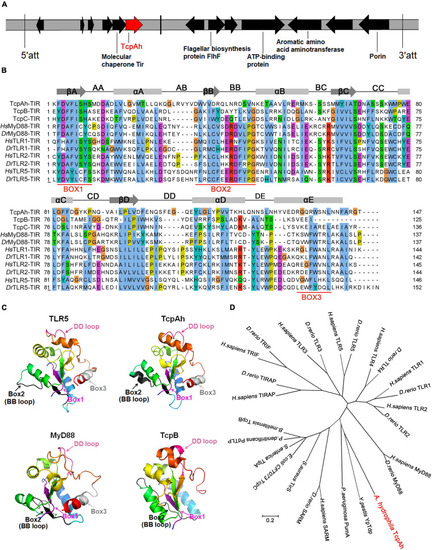
Bioinformatic characterization of TcpAh-encoding gene and protein in A. hydrophila JBN2301 strain. (A) Localization of TcpAh-encoding gene in JBN2301 genome. The encoding sequence of TcpAh is indicated in red arrow and att means attachment site. (B) Sequence alignment of TIR domains using Jalview/ClustalW and ClustalX programs. Conserved functional motifs and amino acid residues are shaded in different colors. Secondary structures were predicted by Jalview/Jpred program. Gray boxes, gray arrows, and black lines denote alpha helices, beta-sheet areas, and connecting loops, respectively. (C) Prediction of the 3D structures of TIR domains using Phyre server and PyMol program. The TcpAh TIR domain (38–184 aa) was modeled with 100% confidence by the single highest scoring template as the TIR domain of TLR5 (PDB c3j0aA). Box 1 (purple), box 2 and BB loop (black), and box 3 and DD loop (hot pink) are indicated within the TIR domains. (D) Phylogenetic trees of the amino acid sequences of TcpAh and its homologs in other species constructed by neighbor-joining method using MEGA 7.0. The tree was drawn to scale, and branch lengths are in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the p-distance method. The GenBank accession numbers of the sequences are as follows: H. sapiens MyD88, AAC50954.1; D. rerio MyD88, AAQ91324.1; H. sapiens TLR1, NP_891549.1; D. rerio TLR1, AAI63271.1; H. sapiens TLR2, AAY85648.1; D. rerio TLR2, NP_997977.1; H. sapiens TLR3, NP_003256.1; D. rerio TLR3, AAI07956.1; H. sapiens TLR4, NP_003257.1; D. rerio TLR4, NP_001315534.1; H. sapiens TLR5, NP_003259.2; D. rerio TLR5, NP_001124067.2; H. sapiens TRIF, AAH09860.2; D. rerio TRIF, NP_001038224.1; H. sapiens TRIAP, NP_001034750.1; D. rerio TRIAP, XP_002667158.2; H. sapiens SARM, NP_055892.2; D. rerio SARM, NP_001124068.1; Y. pestis YpTdp, WP_198249346.1; A. hydrophila TcpAh, WP_043158960; P. aeruginosa PumA, WP_012075302.1; S. aureus TirS, WP_000114516.1; E. coli CFT073 TcpC, WP_000282336.1; S. enterica TlpA, WP_079786625.1; P. denitrificans PdTLP, QAR27511.1; B. melitensis TcpB, EPZ76643.1. PHENOTYPE:
|
|
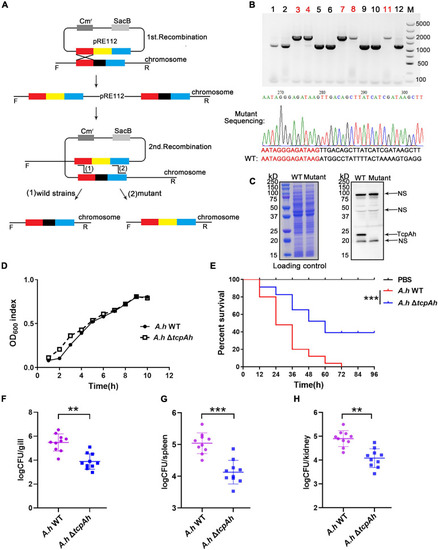
Examination of the requirement of TcpAh for |
|
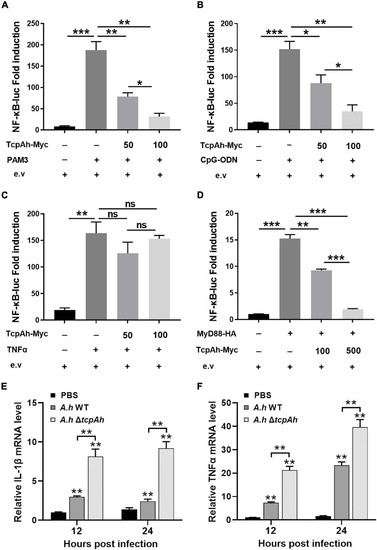
Examination of the inhibitory role of TcpAh in MyD88 signaling pathway. |
|
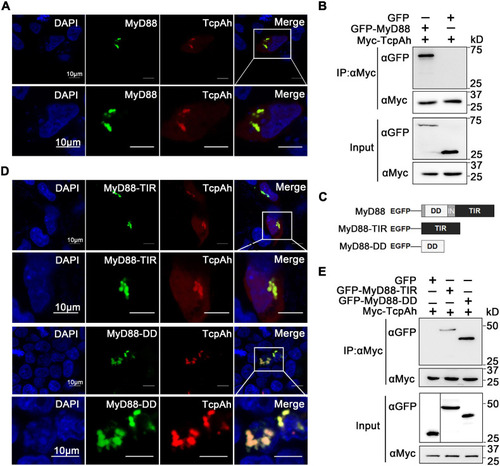
Interaction between TcpAh and MyD88 by TIR–TIR and TIR–DD interactions. |
|
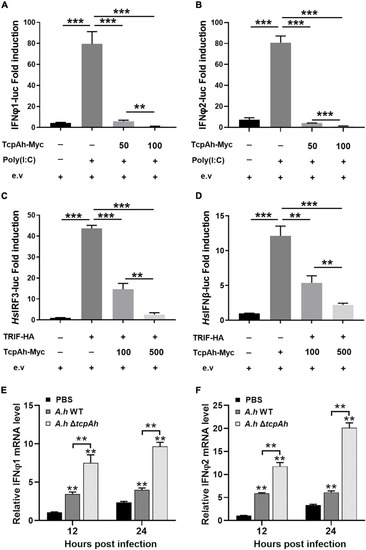
Examination of the inhibitory role of TcpAh in TRIF signaling pathway. |
|
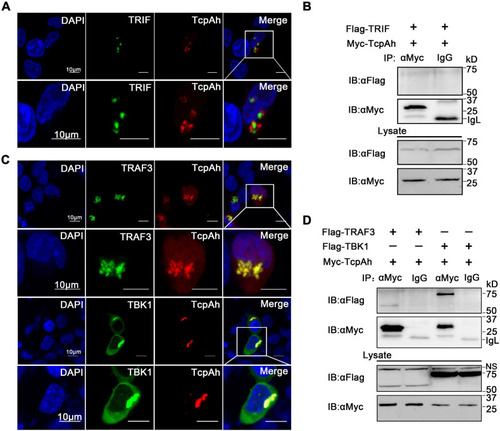
Examination of the associations of TcpAh with TRAF3 and TBK1. |
|
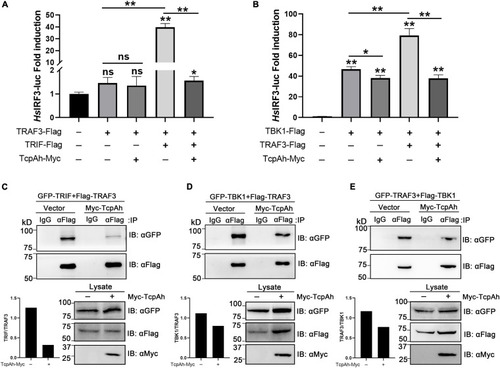
Examination of the functional role of TcpAh in preventing TRIF–TRAF3 and TRAF3–TBK1 interactions. |
|
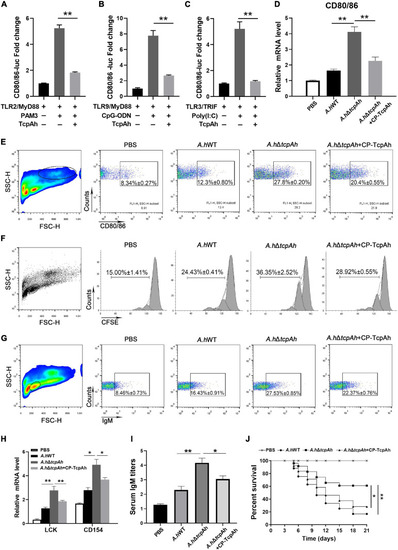
Examination of the inhibitory role of TcpAh in zebrafish adaptive humoral immunity against infection by repressing CD80/86 expression. (A–C) Activation of zebrafish CD80/86 promoter in HEK293T cells transfected with CD80/86 luciferase reporter (CD80/86-Luc; 200 ng/mL), renilla luciferase reporter (15 ng/mL) and expression vectors for zebrafish TLR2 or TLR9 or TLR3 (20 ng/mL) in combination with MyD88 or TRIF (20 ng/mL) with or without TcpAh expression vector (250 ng/mL). After 24 h, the HEK293T cells were stimulated with PAM3 or CpG-ODN or Poly(I:C) for 12 h. Data are the average luciferase activity ± SD (**p < 0.01; ***p < 0.001). (D) Real-time PCR analysis for the expression of zebrafish cd80/86 in leukocytes, which were sorted from peripheral blood, spleen, and kidney tissues 2 days after i.p. stimulation with PBS, wild-type A. hydrophila, ΔtcpAh mutant, or ΔtcpAh mutant complemented with CP-TcpAh protein. Data are representative of three independent experiments as mean ± SD (*p < 0.05; **p < 0.01). (E) Flow cytometric analysis of CD80/86 expression level on MHC-II+ antigen-presenting cells (APCs) of each in vivo treatment group. Data are representative of three independent experiments as mean ± SD (*p < 0.05; **p < 0.01). (F) Proliferation of lymphocytes determined by CFSE dilution through flow cytometry under the indicated experimental treatment. (G) Proliferation of IgM+ B cells determined by flow cytometry under the indicated experimental treatment. Data are representative of three independent experiments as mean ± SD (*p < 0.05; **p < 0.01). (H) Real-time PCR analysis of the expression levels of zebrafish LCK and CD154 of each in vitro treatment group. Data are representative of three independent experiments as mean ± SD (*p < 0.05; **p < 0.01). (I) Examination of the inhibitory role of TcpAh in IgM production in response to A. hydrophila infection in each treatment group by ELISA. Data are representative of three independent experiments as mean ± SD (n = 20; *p < 0.05; **p < 0.01). (J) Examination of the inhibitory role of TcpAh in zebrafish defense against A. hydrophila infection. Zebrafish were infected with A. hydrophila ΔtcpAh or with wild-type A. hydrophila or with A. hydrophila ΔtcpAh complement with CP-TcpAh. Differences were analyzed using log-rank test (*p < 0.05; **p < 0.01). Group of fish injected with mock PBS was used as a negative control. |