- Title
-
Characterization of a member of the CEACAM protein family as a novel marker of proton pump-rich ionocytes on the zebrafish epidermis
- Authors
- Kowalewski, J., Paris, T., Gonzalez, C., Lelièvre, E., Castaño Valencia, L., Boutrois, M., Augier, C., Lutfalla, G., Yatime, L.
- Source
- Full text @ PLoS One
|
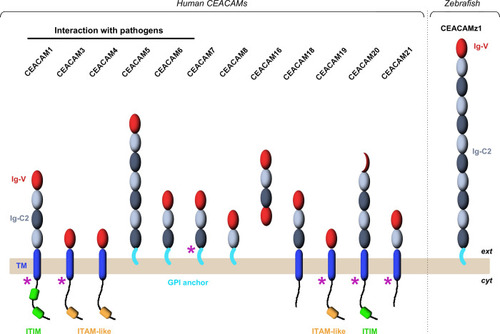
Immunoglobulin domains composing the CEACAM ectodomains are displayed as oval spheres (in red for Ig-V domains; in light or dark grey for A-type or B-type Ig-C2 domains, respectively). Single-pass transmembrane helices are represented by blue, rounded rectangles. GPI anchors are shown in cyan. Cytoplasmic ITIM (Immunoreceptor Tyrosine-based Inhibitory Motif) and ITAM-like (Immunoreceptor Tyrosine-based Activation Motif) motifs are highlighted in green and orange, respectively. Purple arrows indicate the CEACAMs for which several isoforms have been characterized. |
|
|
|
|
|
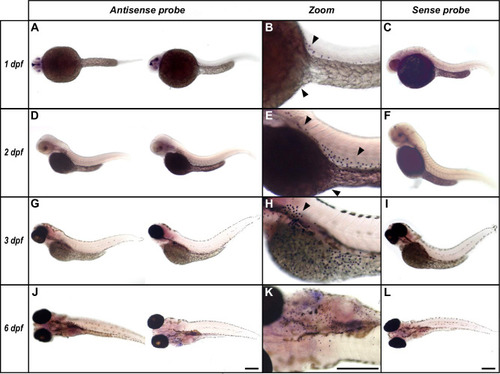
Larvae at 1 dpf |
|
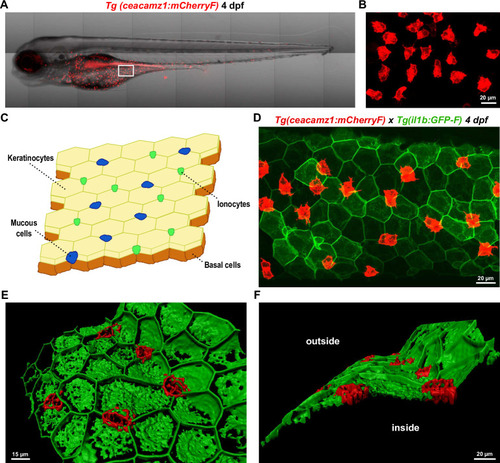
A. Live imaging of a 4 dpf Tg(ceacamz1:mCherry-F) zebrafish larva using spinning disk confocal microscopy. B. Zoom on the ventral region of the 4 dpf transgenic larva as indicated in panel A. C. Schematic representation of the cell composition of zebrafish skin epidermis at early larval stages. D. Z-projection of a portion of the ventral skin of a 4 dpf double-crossed Tg(ceacamz1:mCherry-F) x Tg(il1b:eGFP-F) zebrafish larva. CEACAMz1-expressing cells intercalate within the pavement of keratinocytes. E. and F. 3D-reconstructions of the Z-projection shown in panel D using IMARIS. Top view (E) and lateral view (F). CEACAMz1-expressing cells are embedded within the same layer as keratinocytes and they protrude on both sides of this layer. EXPRESSION / LABELING:
|
|
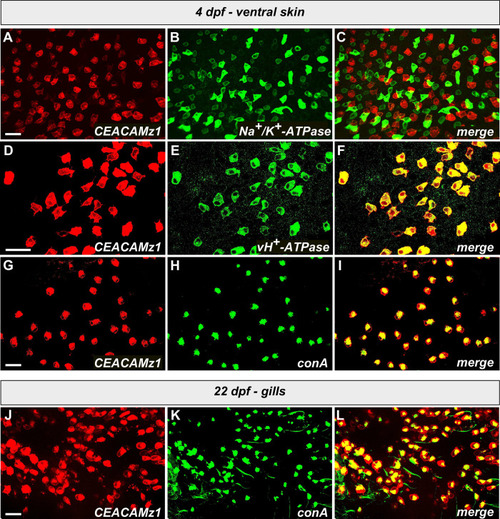
EXPRESSION / LABELING:
|
|
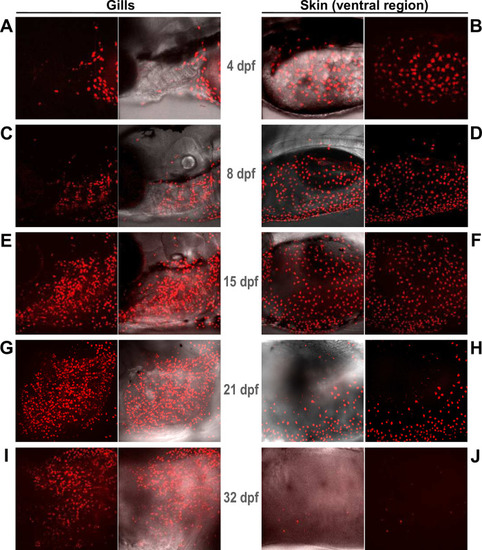
Live imaging of larvae from the EXPRESSION / LABELING:
|
|
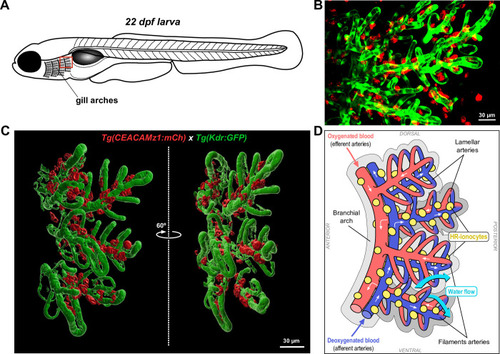
EXPRESSION / LABELING:
|