- Title
-
Correlative microscopy and block-face imaging (CoMBI) method for both paraffin-embedded and frozen specimens
- Authors
- Ishii, N., Tajika, Y., Murakami, T., Galipon, J., Shirahata, H., Mukai, R., Uehara, D., Kaneko, R., Yamazaki, Y., Yoshimoto, Y., Iwasaki, H.
- Source
- Full text @ Sci. Rep.
|
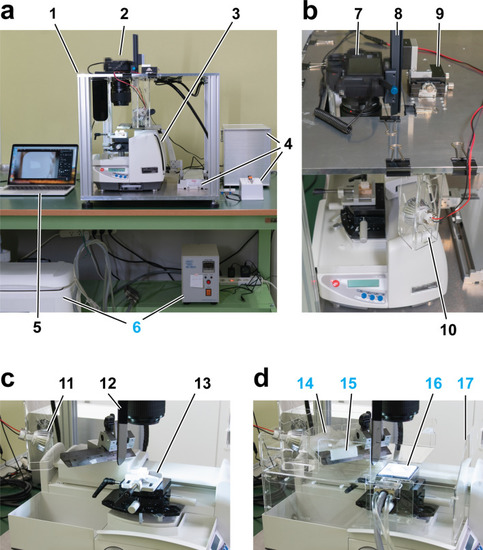
CoMBI system using a sliding microtome. CoMBI-S. ( |
|
Correlation between 3D image and sections of juvenile zebrafish. A juvenile zebrafish (30 dpf) is sliced at a thickness of 5 µm using CoMBI-S. ( |
|
Opacification of paraffin blocks using white agarose gel. Specimens are pre-embedded in agarose gel containing white watercolor, then dehydrated and embedded in paraffin. ( |
|
Opacification of paraffin block using white crayon. (s) A piece of broccoli was embedded in paraffin containing 6.25% w/w white crayon and placed on the custom-made attachment (asterisk). (b) The custom-made white attachment was designed (upper image) and made using 3D printing machine (lower image). Characteristics of the attachment are the white color and the sawtooth pattern on the top, which avoid shadows and increase the area for attaching paraffin block. (c) The border between broccoli specimen and white paraffin is apparent on the block face (upper image). When the block-face image was converted to grayscale image for volume rendering, only the cutting face of the broccoli specimen is seen (lower image). (d) The serial block-face images (270 images with a voxel size of 20 × 20 × 20 µm) were converted to grayscale images (c), then used for volume rendering (d). Bars: 5 mm. The STL file of attachments is available at GitHub |
|
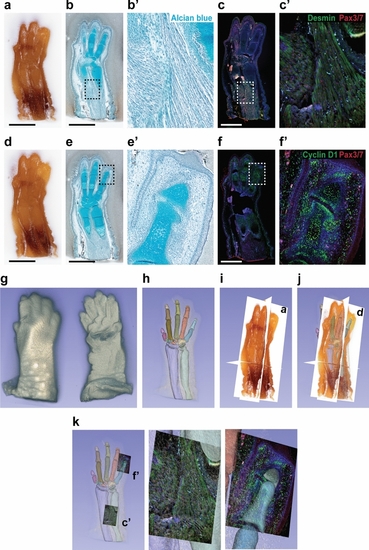
Usability of CoMBI data. Mouse forelimb on E16 were pre-embedded in white-agarose and embedded in paraffin. The 267-serial block-face images with 6-µm interval were obtained. ( |
|
Correlation between 3D image and sections of mouse embryo on E10 in a frozen block. The mouse embryo on E10 was frozen and sliced at a thickness of 10 µm using CoMBI-S system. ( |
|
Tannic acid staining improves the visibility of tissues in the frozen blocks. ( |
|
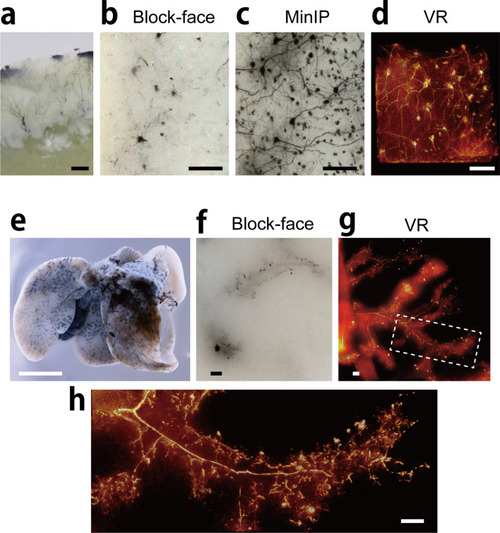
Fine 3D imaging at cellular level. ( |