- Title
-
Association Analysis of Variants of DSCAM and BACE2 With Hirschsprung Disease Susceptibility in Han Chinese and Functional Evaluation in Zebrafish
- Authors
- Lu, Y.J., Yu, W.W., Cui, M.M., Yu, X.X., Song, H.L., Bai, M.R., Wu, W.J., Gu, B.L., Wang, J., Cai, W., Chu, X.
- Source
- Full text @ Front Cell Dev Biol
|
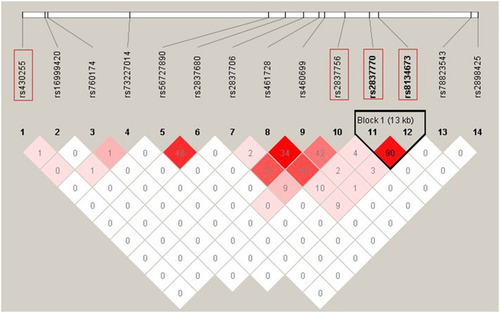
Linkage disequilibrium (LD) pattern of 12 |
|
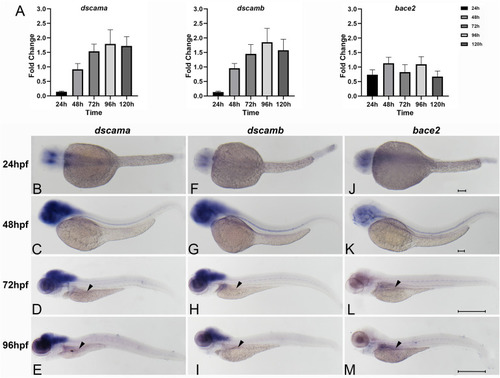
Spatiotemporal expression of zebrafish of |
|
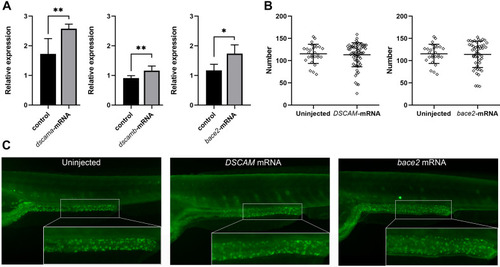
Phenotypes of zebrafish embryos with overexpression of |
|
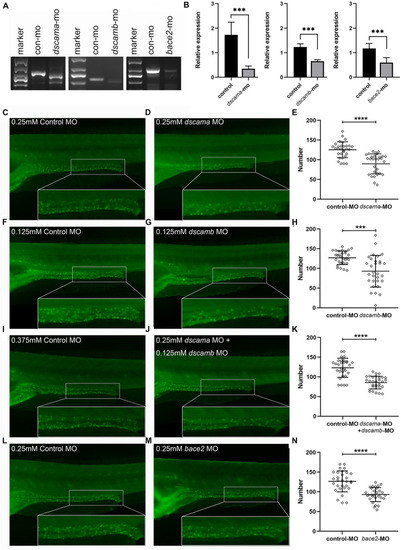
Phenotypes of PHENOTYPE:
|
|
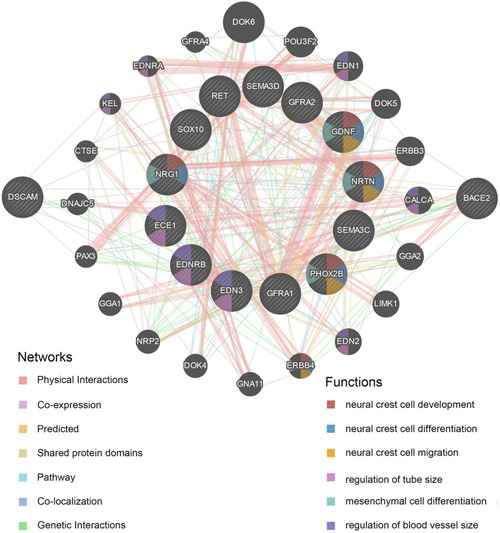
Protein–protein interaction (PPI) network of |