- Title
-
Abortive intussusceptive angiogenesis causes multi-cavernous vascular malformations
- Authors
- Li, W., Tran, V., Shaked, I., Xue, B., Moore, T., Lightle, R., Kleinfeld, D., Awad, I.A., Ginsberg, M.H.
- Source
- Full text @ Elife
|
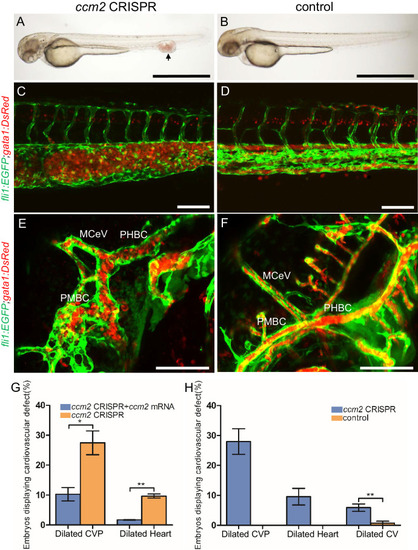
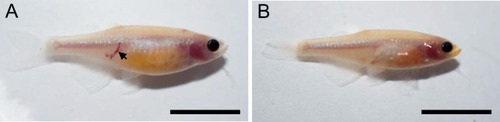
Endothelial cells and red blood cells were labeled by EGFP and DsRed respectively in double transgenic Tg(fli1:EGFP)y1;Tg(gata1:DsRed)sd2 embryos. (A) Red blood cells accumulate in dilated segments of the caudal vein of ccm2 CRISPR fish at 2 days post fertilization (dpf). (B) cas9 mRNA-injected control embryo. (C) ccm2 CRISPR embryos showed accumulation of red blood cells and intraluminal endothelial cells in a dilated segment of caudal vein in contrast to a control embryo. Note: In this and all succeeding sagittal views, anterior is to the left (D). (E) ccm2 CRISPR embryos occasionally showed dilations of cerebral veins, whereas control embryos (F) showed normal development of cerebral veins (F). MCeV: mid-cerebral vein, PMBC: primordial midbrain channel, PHBC: primordial hindbrain channel. (G) The dilated caudal venous plexus (CVP) and heart of ccm2 CRISPR embryos were rescued by ccm2 mRNA injection. p=0.0336 (dilated CVP), 0.0037 (dilated heart). p-Values were calculated using an unpaired two-tailed Student’s t-test. (H) Phenotypic distribution of dilated heart, CVP, and cerebral veins (CV) in ccm2 CRISPR embryos at 2 dpf. p=0.0078 (dilated CVP), 0.0268 (dilated heart), 0.0041 (dilated CV). p-Values were calculated using a paired two-tailed Student’s t-test. Error bars indicate SD. Scale bar: 1 mm in A and B, and 100 µm in C through F. PHENOTYPE:
|
|
( |
|
Microinjection was performed on PHENOTYPE:
|
|
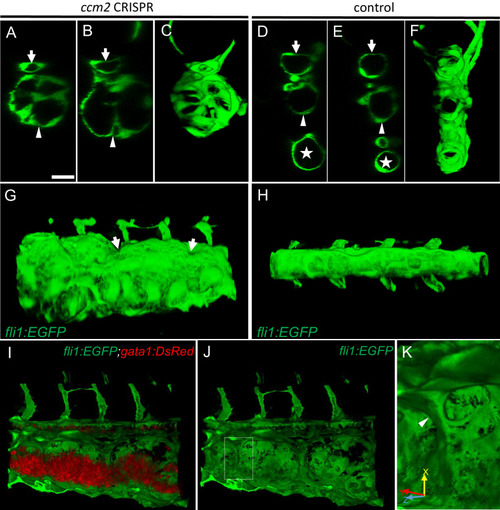
(A–F) XZ planes and three-dimensional (3D) projection along Y axis of Airyscan images revealed intraluminal endothelial pillars at 2 days post fertilization (dpf) (A–C), whereas Cas9-injected control embryos displayed a normal patent lumen in both a dorsal and ventral caudal vein (D–F). Endothelial cells were labeled by EGFP in Tg(fli1:EGFP) embryos. Arrow, arrowhead, and asterisk indicated the dorsal aorta, dorsal vein, and ventral vein, respectively. (G and H) Ventral view of 3D reconstruction show the irregular surface of the dramatically dilated caudal vein segment in ccm2 CRISPR embryo (G) and normal ventral vein (H). Arrows in G indicate small pits where the endothelial pillars originate. (I–K) Intraluminal view of 3D reconstruction of ccm2 CRISPR embryo reveals the intraluminal pillars honeycombing the lumen and the accumulated red blood cells (I). Erythrocytes were not imaged in J to reveal pillars and the area within the box in (J) was magnified in (K), and arrowhead indicates the intravascular pillar. Endothelial cells and red blood cells were labeled by EGFP or DsRed respectively in Tg(fli1:EGFP)y1;Tg(gata1:DsRed)sd2 embryos. Scale bar: 20 µm. PHENOTYPE:
|
|
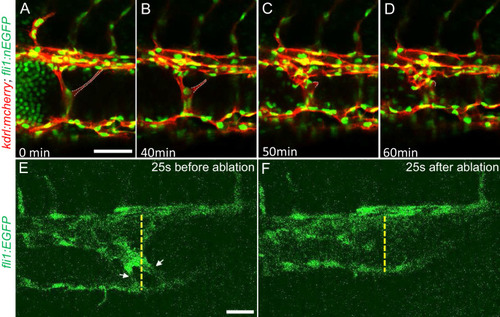
(A through D) Time lapse images reveal spontaneous retraction of an intravascular pillar leading to re-entry of blood cells into circulation and reduced dilation of the caudal vein. Endothelial cells were labeled by mCherry, and their nucleus and some red blood cells were labeled by EGFP in the Tg(fli1:nEGFP)y7;Tg(kdrl:mcherryras)s896 embryos. The retracted pillar is outlined by dotted lines for emphasis. Note that pillar retraction and vessel dilation were temporally correlated. (E and F) Laser ablation of pillar reduced caudal venous plexus (CVP) diameter. The diameter of the dilated vein (E) was reduced after ablation (F). Note the pillars indicated by arrows in (E) are gone after ablation in (F). Dashed line indicates the diameter of the vein before and after ablation. Scale bar: 50 µm. |
|
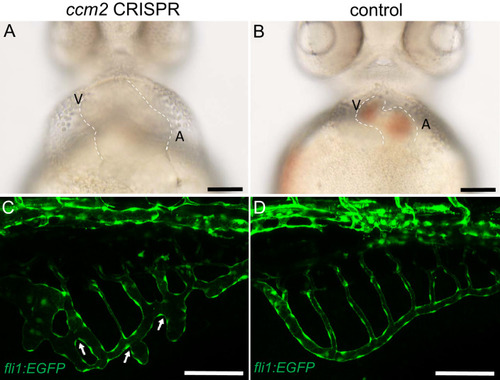
A PHENOTYPE:
|
|
Morpholinos targeting tnnt, gata1, tif1gamma, or a control morpholino were co-injected with ccm2 guide and Cas9 RNA. (A) Reduction of blood flow in tnnt morphants (A, B, C) resulted in reduced caudal venous plexus (CVP) dilation (A) and intravascular honeycombing (B, C) in 2 days post fertilization (dpf) ccm2 CRISPR Tg(fli1:EGFP) embryos. Arrows indicate intussusceptions. Scale bar: 100 µm. (A) Loss of erythrocytes in gata1 or tif1gamma morphant ccm2 CRISPR embryos also reduced the incidence of CVP dilation. p-Values were calculated using one-way ANOVA. **p<0.01. Error bars indicate SD. (D and E) At 23 hpf, ccm2 CRISPR Tg(klf2a:H2b-EGFP) embryos displayed a mosaic increase of EGFP expression in endothelial cells in the CVP (D), compared with cas9 mRNA control embryos (E). Scale bar: 25 µm. (F) Quantification of the EGFP fluorescence intensity using ImageJ. A total of 20 nuclei were analyzed from ccm2 CRISPR embryos, and 16 nuclei were analyzed from control embryos. Note that 11 nuclei in CRISPR embryo displayed intensity above 3000, while all of the nuclei in control embryo are below 3000. (G) ccm2 CRISPR and tnnt morpholino-injected Tg(klf2a:H2b:EGFP 2 dpf) embryos displayed a mosaic increase of endothelial nuclear EGFP expression in dorsal vein. Scale bar: 50 µm. In A through C, EGFP expression was driven by klf2a promoter in Tg(klf2a:H2b:EGFP) embryo, and endothelial cells were labeled by mcherry in Tg(kdrl:mcherry) transgenic line. Arrows indicated the endothelial nuclei with increased EGFP, and arrowheads indicated the other endothelial nuclei along the ventral wall of dorsal vein. EXPRESSION / LABELING:
PHENOTYPE:
|
|
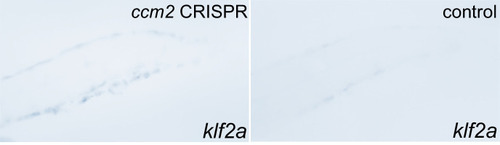
Whole mount in situ hybridization showed mosaic upregulation of klf2a expression in |
|
( |
|
(A) Both the CVP dilation and heart dilation were rescued by injection of klf2 morpholinos in 2 days post fertilization (dpf) ccm2 CRISPR embryos. **p<0.01. Error bars indicate SD. (B) pCS2-KLF2a linearized DNA-injected 2.5 dpf embryos displayed CVP dilation, whereas injection of a DNA fragment containing a DNA binding domain deleted ΔKLF2a mutant showed normal development. Arrow indicates the CVP dilation and retained erythrocytes. Scale bar: 1 mm. (C) Quantification of the prevalence of CVP dilation following KLF2a or ΔKLF2a overexpression. The mean and SD are shown. (D) Representative images show the honeycombed lumen and dilated CVP in 1.5 dpf KLF2a-injected embryo and normal CVP of ΔKLF2a-injected embryo. Arrow indicates honeycombing. Scale bar: 100 µm. PHENOTYPE:
|
|
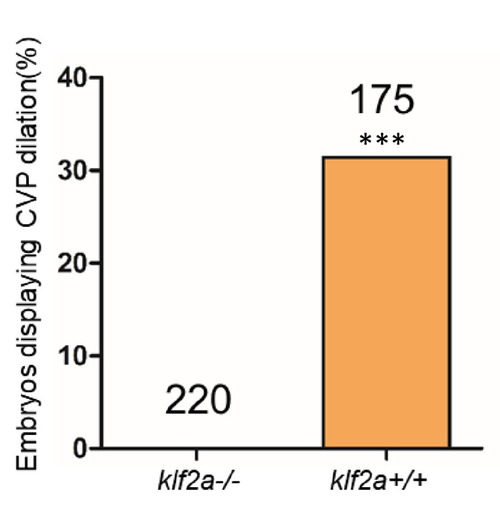
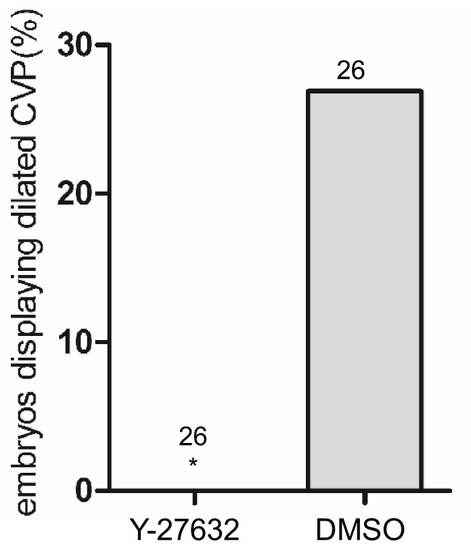
Total number of embryos in each group is indicated on the graph. ***p<0.0001. Two-tailed Fisher’s exact test was used for comparisons. |
|
(A) Low-dose ccm2 morpholino reduced the incidence of CVP dilation but did not significantly increase heart dilation in ccm2 CRISPR embryos. (B and C) Mosaic ccm2 but not inactive ccm2(L197E) overexpression caused CVP dilation. Arrows indicate pillars in the CVP. (B’ and C’) Mosaic expression of mOrange-tagged ccm2 or ccm2(L197E). Scale bar: 100 µm. Error bars are ± SD. PHENOTYPE:
|
|
The 200 ng/μl linearized DNA fragment containing CMV promoter, |
|
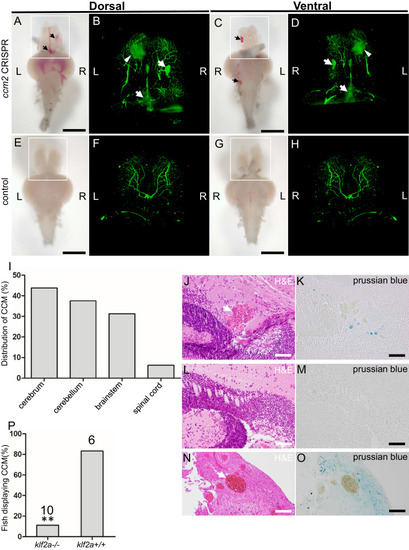
The ~50% of ccm2 CRISPR fish that survived developed highly penetrant CCMs (A and C). Arrows indicate superficial lesions on dorsal (A) and ventral (C) surface of the brain. Note hemorrhage into the ventricles. Lesions are absent in control embryos (E and G). Clear, unobstructed brain imaging cocktails and computational analysis (CUBIC) clearing (B, D, F, H) enables visualization of CCM burden by light sheet microscopy. Arrows indicate the lesions that corresponded to those seen in bright field, and arrowhead indicates a deeper lesion. L: left, R: right. Scale bar: 1 mm. (I) Cavernomas were dispersed throughout the central nervous system including cerebrum, cerebellum, brain stem, and spinal cord. (J) Hematoxylin and eosin (H&E) stained brain section reveals nucleated erythrocytes filling a dilated vessel with adjacent Prussian blue stained iron deposition (K) in ccm2 CRISPR fish and the absence of lesions or iron deposition in control fish (L, M). (N, O) A CCM from a patient stained with H&E (N) or Prussian blue (O). Note similar appearance to the zebrafish lesion shown in (J, K). Arrow indicates dilated vessel. Scale bar: 50 µm. (P) CCMs were significantly reduced in ccm2 CRISPR adult fish on klf2a-/- background compared to that on klf2a+/+ background. Total number of embryos in each group is indicated. p=0.0076. Two-tailed Fisher’s exact test was used for comparison. PHENOTYPE:
|
|
Casper fish embryos were injected with |
|
At 24 hpf, PHENOTYPE:
|