- Title
-
How Zebrafish Can Drive the Future of Genetic-based Hearing and Balance Research
- Authors
- Sheets, L., Holmgren, M., Kindt, K.S.
- Source
- Full text @ J. Assoc. Res. Otolaryngol.
|
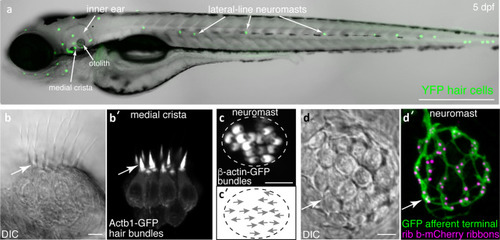
Visualization of structures in hair cell systems of larval zebrafish. |
|
Outline of mutagenesis in zebrafish used for TILLING and forward genetic screening. Both retroviruses and the mutagen ENU can be used to create germline mutations in zebrafish. For retroviral-based mutagenesis, a DNA construct containing the retrovirus is injected into newly fertilized zebrafish embryos. These injected embryos are grown to adulthood (2–3 months), resulting in G0 adults that are mosaic for germline mutations. G0 adults are then outcrossed to wildtype adults to generate G1 adults that are heterozygous for different genetic lesions. In chemical mutagenesis, adult males are treated with a chemical mutagen such as ENU. These mutagenized males are crossed to wildtype adult females to generate G1 adults that are heterozygous for different genetic lesions. For a forward genetic screen (ENU or retroviral-based mutagens), G1 adults are crossed to wildtype animals, providing a pool of G2 adult carriers. G2 adults are incrossed, and G3 larvae are screened for a phenotype of interest. When mutagens are used to screen for a specific genetic lesion (in the case of TILLING), G1 adults or sperm stored in a library from G1 males are screened for mutations. After identifying a specific mutation, the identified G1 adult is crossed to wildtype to generate G2 adults harboring that mutation. Two G2 adults with the lesion of interest are then incrossed and screened for phenotypes. Blue and green represent distinct genetic lesions in mutagenized fish |
|
GFP-based forward genetic screen reveals mutants with swollen afferent terminals. ( |
|
Outline of Morpholino timeline in zebrafish. Morpholinos (MOs) are used to block mRNA splicing or translation of a particular gene product. MOs are injected into newly fertilized zebrafish embryos. Two to 5 days after MO injection morphant larvae can be screened for phenotype. The penetrance of MO phenotypes is highly variable. It is recommended that MO phenotypes be confirmed using a germline mutant when possible (Stainier et al. |
|
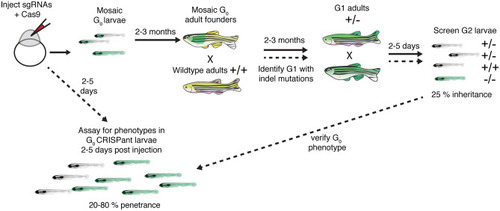
Outline of how to create germline zebrafish CRISPR-Cas9 mutants and CRISPants. To create a germline zebrafish CRISPR-Cas9 mutant (follow path of solid lines), guide RNAs (gRNAs) targeting a gene of interest along with Cas9 mRNA or protein are injected into newly fertilized zebrafish embryos. These G0 injected embryos are grown to adulthood (2–3 months). G0 adult founders are crossed to wildtype adults, and the G1 progeny screened for indels. G1 progeny with indels are grown to adulthood. G1 adults containing indels are then incrossed and screened for phenotypes. To perform analyses on injected G0 CRISPant larvae, optimized gRNAs are injected into newly fertilized zebrafish embryos. Injected CRISPant G0 larvae are then screened for phenotypes days after the injection. CRISPant phenotypes can be verified by generating a stable, germline mutant |
|
Two examples G0 |
|
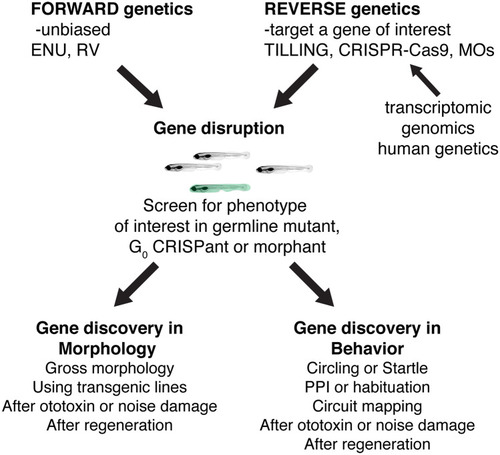
Past, present, and future ways to use zebrafish genetics to study hearing and balance. Both forward and reverse genetic approaches in zebrafish have had an immense impact on gene discovery in hearing and balance. In the future, novel forward genetic screens using transgenic lines or novel damage paradigms have the potential to continue this path to gene discovery. In addition, advances in reverse genetics will continue to provide a valuable way to screen genes implicated in humans hearing loss. Reverse genetic screening many also prove a valuable, high-throughput pipeline to validate transcriptomics or genomics datasets |