- Title
-
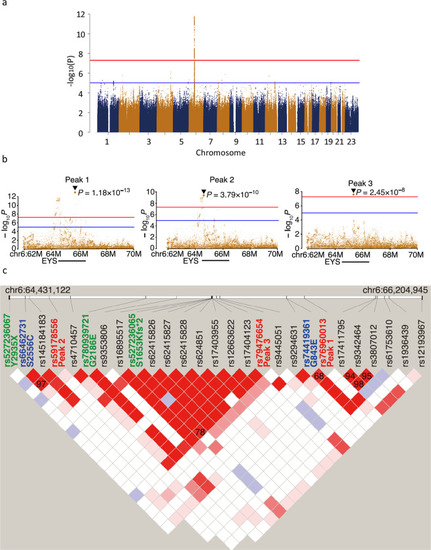
A hypomorphic variant in EYS detected by genome-wide association study contributes toward retinitis pigmentosa
- Authors
- Nishiguchi, K.M., Miya, F., Mori, Y., Fujita, K., Akiyama, M., Kamatani, T., Koyanagi, Y., Sato, K., Takigawa, T., Ueno, S., Tsugita, M., Kunikata, H., Cisarova, K., Nishino, J., Murakami, A., Abe, T., Momozawa, Y., Terasaki, H., Wada, Y., Sonoda, K.H., Rivolta, C., Tsunoda, T., Tsujikawa, M., Ikeda, Y., Nakazawa, T.
- Source
- Full text @ Commun Biol
|
|
|
|
|
a Domain structure of EYS in relation to G843E. b Conservation of G843 across diverse species ranging from zebrafish to humans. The multiple sequence alignment was generated using ClustalW. Accession numbers of the protein sequences used for sequence comparison are as follows: human, NM_001142800.1; macaque, XM_011737495.1; pig, XM_021084496; chicken, XM_015284845.1; zebrafish, XM_009307513. |
|
|