- Title
-
Extensive nuclear gyration and pervasive non-genic transcription during primordial germ cell development in zebrafish
- Authors
- Redl, S., de Jesus Domingues, A.M., Caspani, E., Möckel, S., Salvenmoser, W., Mendez-Lago, M., Ketting, R.F.
- Source
- Full text @ Development
|
|
|
|
|
|
|
|
|
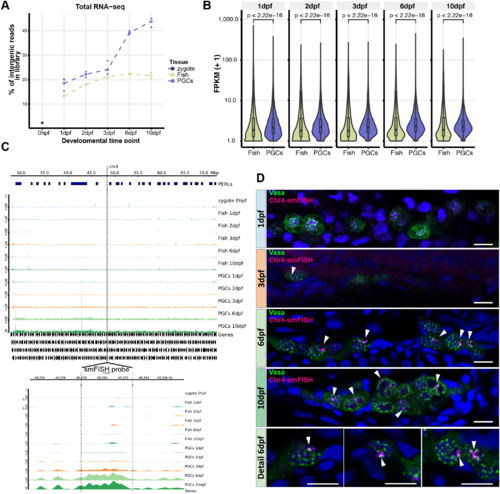
Expression of intergenic regions in the PGCs. (A) Proportion of intergenic reads in total RNAseq libraries at the different indicated time points. (B) Distributions of the expression levels of intergenic regions at the different time points in fragments (+1) per kilobase of transcript per million mapped reads (FPKM). Significance of the differences between ‘Fish’ and ‘PGC’ samples was tested using a Mann–Whitney-Wilcoxon test. (C) Examples of the right of chromosome 4 as a region with many PERLs. The region targeted for smFISH is indicated with a dashed line and enlarged in the bottom panel. It does not overlap with the PERLs, as annotated by our scripts, but still shows a clear increase in expression. (D) Immunostaining for Vasa, combined with smFISH using a probe designed to target transcripts derived from chr4: 48256970-48262902. White arrowheads indicate smFISH signal. Scale bars: 10 µm. |
|
|
|
|