- Title
-
Automated high-throughput heartbeat quantification in medaka and zebrafish embryos under physiological conditions
- Authors
- Gierten, J., Pylatiuk, C., Hammouda, O.T., Schock, C., Stegmaier, J., Wittbrodt, J., Gehrig, J., Loosli, F.
- Source
- Full text @ Sci. Rep.
|
Workflow of automated imaging and heart rate quantification in medaka and zebrafish embryos. ( |
|
Functionalities of |
|
Heartbeat detection of multiple embryos per well. ( |
|
Heart rates of medaka and zebrafish during embryonic development. ( |
|
Temperature-dependent heart rate modulation. ( |
|
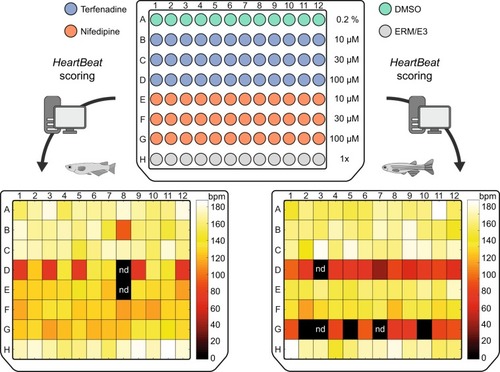
Quantification of heart rates in fish embryos treated with compounds. 96-well format for incubation of medaka and zebrafish embryos with terfenadine and nifedipine each at three different concentrations in µM. As a control for stage-dependent changes of heart rate, all measurements (drugs and DMSO) were normalized to a negative control group of age-matched untreated embryos (row H). The |
|
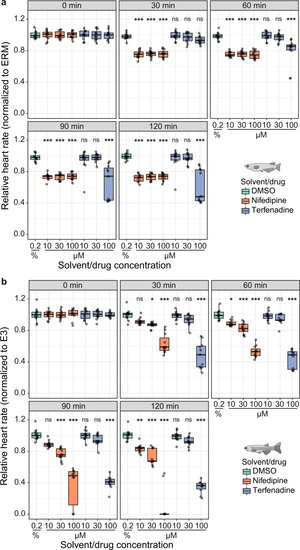
Heart rate inhibition by nifedipine and terfenadine over time. ( |