- Title
-
SPRTN protease and checkpoint kinase 1 cross-activation loop safeguards DNA replication
- Authors
- Halder, S., Torrecilla, I., Burkhalter, M.D., Popović, M., Fielden, J., Vaz, B., Oehler, J., Pilger, D., Lessel, D., Wiseman, K., Singh, A.N., Vendrell, I., Fischer, R., Philipp, M., Ramadan, K.
- Source
- Full text @ Nat. Commun.
|
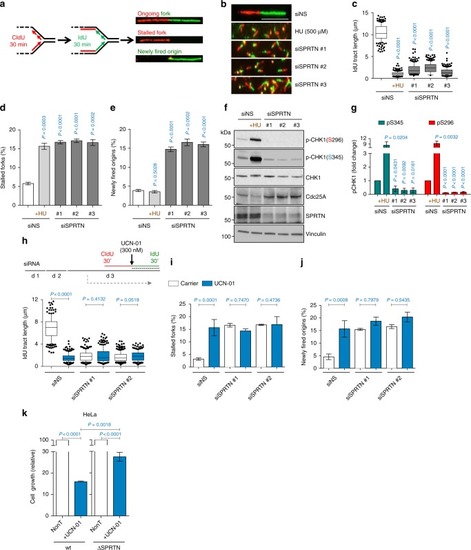
SPRTN depleted cells endure severe DNA replication stress but fail to activate a CHK1 response. |
|
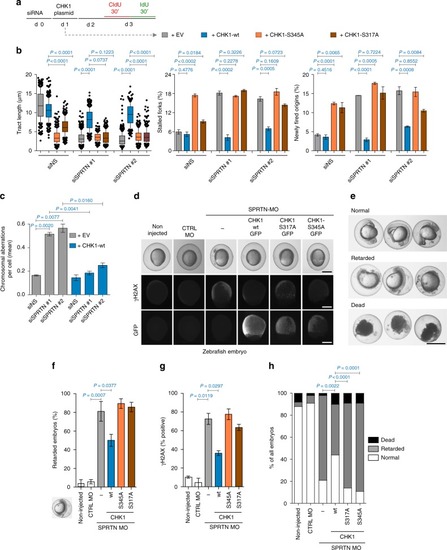
Ectopic CHK1 overexpression rescues SPRTN-deficiency phenotypes. PHENOTYPE:
|
|
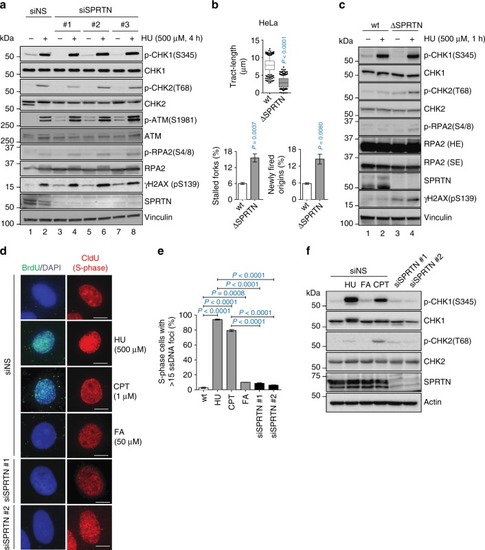
SPRTN does not contribute to the activation of CHK1 after DSB formation. |
|
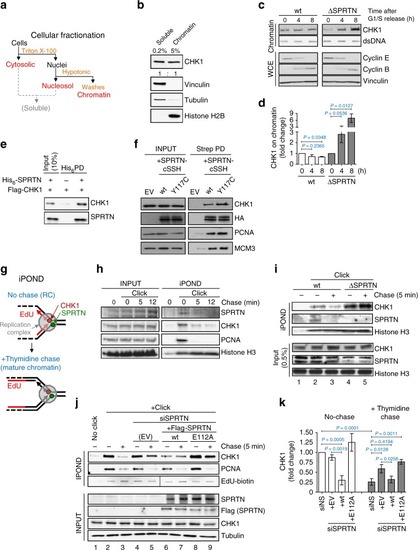
SPRTN protease evicts CHK1 from replicative chromatin. |
|
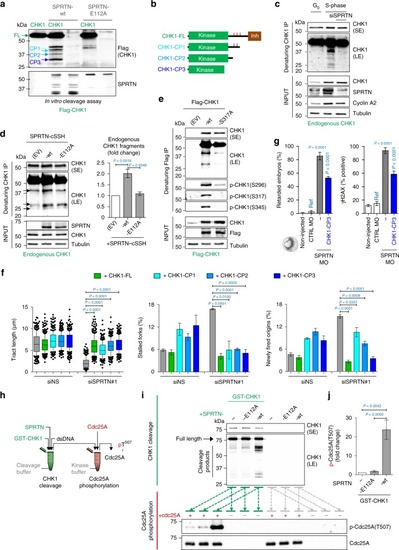
SPRTN cleaves CHK1 and releases kinase-active CHK1 fragments. PHENOTYPE:
|
|
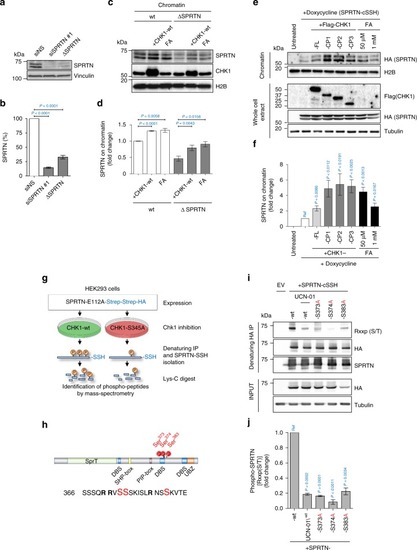
CHK1 phosphorylates SPRTN and regulates its recruitment to chromatin. |
|
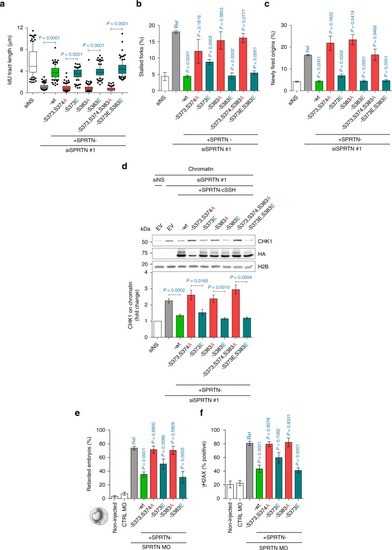
CHK1 targeted phosphorylation of SPRTN is essential for its biological function. PHENOTYPE:
|
|
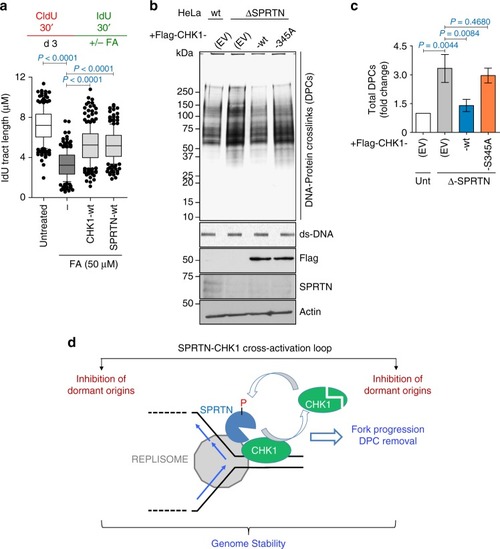
CHK1 and SPRTN promote replication fork progression by removing DPCs. |