- Title
-
Discovering the chloride pathway in the CFTR channel
- Authors
- Farkas, B., Tordai, H., Padányi, R., Tordai, A., Gera, J., Paragi, G., Hegedűs, T.
- Source
- Full text @ Cell. Mol. Life Sci.
|
Channels with extracellular and intracellular ends. Tunnels suitable for chloride conductance were identified in one case out of 22 equilibrium simulations with the ATP-bound CFTR structure (PDBID: 5W81) using Caver. The channels with top scores (spheres with red colors) were clustered based on the atom positions of the channel-lining residues. Intracellular residues with positive charges at the TM4/6 entry site are labeled with blue sticks. Lasso/L0 region, TMD1, TMD2, NBD1, and NBD2 domains are marked with dark green, pale green, green, yellow, and orange, respectively. Lipid molecules lining the pore in the region of TM8 are shown by black sticks. |
|
Comparing channel-lining residues identified in simulations to experimental data. Residues interacting with the Caver spheres were counted from all open frames ( |
|
The chloride channel profile. |
|
Interaction sites of chloride ions with the CFTR protein. Contact sites are depicted by stick representation of amino acids and color-coded according to the contact frequency from green (low) to red (high). |
|
Chloride ions entered the pathway but did not pass. |
|
Coupling helix 4 dynamics is strongly coupled to the bottleneck region. Optimal and suboptimal paths in the network of amino acids (spheres), connected by edges (lines) corresponding to correlation in motions, were determined. Number of suboptimal paths between coupling helix 4 (CH4, L1065) and R334 ( |
|
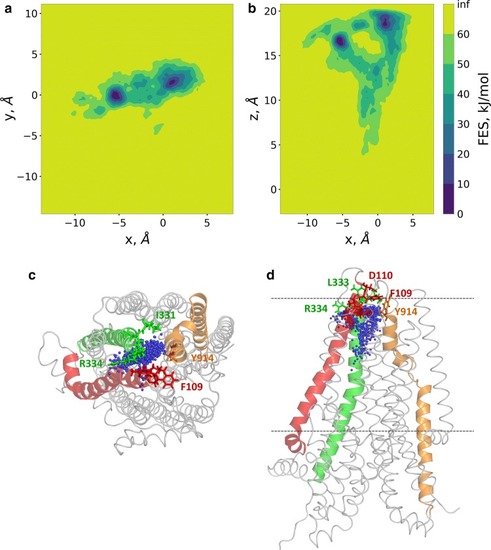
Two routes towards the extracellular regions were detected by metadyanamics simulations. Well-tempered metadynamics simulations were performed to identify potential exit routes of chloride ions and the amino acids narrowing this region of the pathway. The distance between the center of mass of four Cα atoms (a.a. 95, 347, 924, and 1141) and Cl− #2 was used as a reaction coordinate (collective variable, CV). 2D free energy surfaces were calculated along the |