- Title
-
Circadian clock regulation of the cell cycle in the zebrafish intestine
- Authors
- Peyric, E., Moore, H.A., and Whitmore, D.
- Source
- Full text @ PLoS One
|
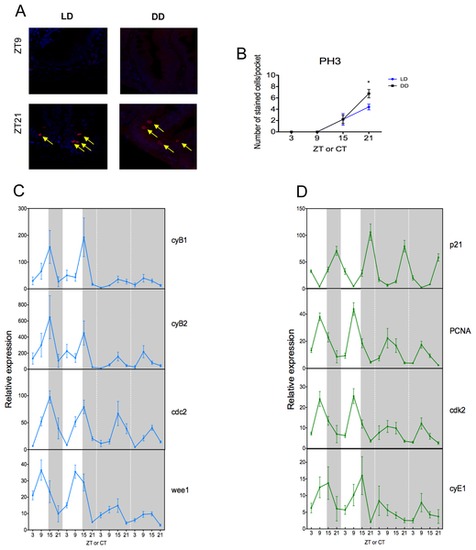
Figure 2. M phase is rhythmic and under circadian control. (A) In LD and DD conditions, cells stained with an anti-pH3 antibody to monitor mitosis (in red and marked by the yellow arrow) at the peak and trough time points, are localized inside the intervillus pockets of the gut. (B) Cell division is rhythmic and is exhibiting a peak of stained cells per pocket at ZT21. This rhythmicity is maintained in DD, showing that cell division is under circadian control. DAPI is used here as a nuclear counterstain. (C–D) Quantitative PCR analysis of endogenous cell cycle genes from M phase (C) and G1/S phase (D). (C) In LD, cyB1, cyB2 and cdc2 are rhythmic and peak at ZT15, whereas wee1 peaks at ZT9. All genes continue to oscillate after entry into DD. (D) In LD, p21, PCNA and cdk2 show a strong rhythm with peak expression at ZT9 for PCNA and cdk2 and a peak at ZT21 for p21. In DD, all genes continue to oscillate robustly with the possible exception of cyE1, which shows quite variable expression even in LD. White and grey backgrounds represent light and dark phases, respectively. Cell cycle gene expression data represents the mean ± SEM from 8 fish per time point. For each time point in panel B, LD data are compared to DD using a Student’s t-test (* p<0.05). All other data are expressed as mean ± SEM. |
|
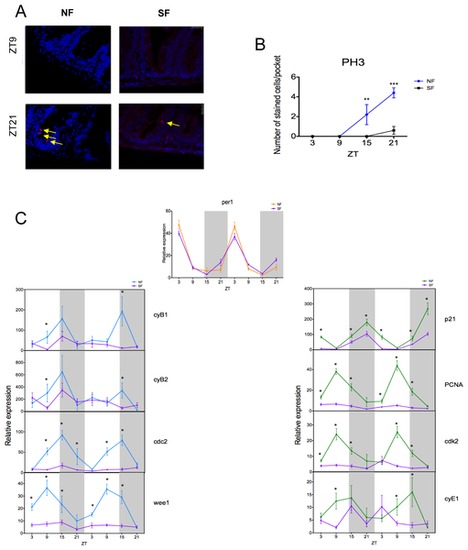
Figure 3. M phase is affected by starvation. (A) Cell division is rhythmic under a normal feeding schedule (NF) (peak and trough shown), but this rhythm is lost when fish are starved (SF). DAPI is used here as a nuclear counterstain. (B) Cell division is largely abolished when no food is given. (C) The per1 rhythm is unaltered in NF and SF fish. However, all the M-phase genes studied (cyB1, cyB2, cdc2 and wee1) and most G1/S-phase genes (PCNA, cdk2 and cyE1) show reduced levels of expression, and a general loss of rhythmicity during starvation. p21 expression is the one exception, showing a relatively small response to starvation. White and grey backgrounds represent light and dark phases, respectively. Cell cycle gene data represents the mean ± SEM from 8 to 12 fish per time point. For panels B and C, NF data are compared to SF using a Student’s t-test (* p<0.05, ** p<0.01 and *** p<0.001). |
|
Mitosis in the zebrafish intestine is rhythmic and under circadian control, but is abolished during starvation. Extended time course of cell division, using a pH3 antibody as a mitotic marker, for fish fed twice a day and entrained to a LD cycle then transferred into DD, as well as for starved fish (SF) entrained to a LD cycle. Gut samples were collected every 6h for the three conditions. In LD, cells divide rhythmically with a peak at ZT21; when the fish are free-running in DD, this rhythmicity is maintained. When the fish are starved, no cell division is observed. |