- Title
-
Systematic human/zebrafish comparative identification of cis-regulatory activity around vertebrate developmental transcription factor genes
- Authors
- Navratilova, P., Fredman, D., Hawkins, T.A., Turner, K., Lenhard, B., and Becker, T.S.
- Source
- Full text @ Dev. Biol.
|
The overview of genomic locus of SOX3. (A) Ancora view of a 2 Mb window across the human locus (upper track), density of HCNEs compared with the mouse genome (lower track) and conserved synteny in the zebrafish genome. (B) 1 Mb window mVISTA plot of human SOX3 compared with zebrafish (dr, upper track), Xenopus tropicalis, (xt, middle track) and Fugu rubripes (fr, lower track). HCNEs conserved in all four species were tested in zebrafish and are boxed in red. The insets above tracks show two enhancer detection insertions in the sox3 gene desert in zebrafish and the RNA in situ hybridization pattern of sox3 in a 2-day zebrafish embryo. Expression is mainly confined to the nervous system (for detailed expression domains see also Supplementary online database). |
|
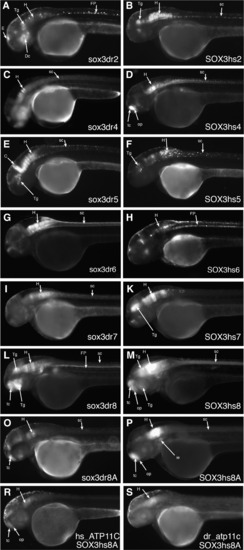
Lateral views of HCNE-test transgenic zebrafish larvae expressing GFP at 2 days post fertilization. Left column shows transgenic made with zebrafish HCNEs, right column shows transgenes made with the orthologous human sequence. (A–M) Elements depicted in Fig. 1, direct expression of GFP to telencephalon (C, D, L–P), diencephalon (A, B, E, F, L–P), hindbrain (A–P) and spinal cord (A–M). (O, P) Partial sequence of element dr8 and hs8; (R, S) Transgenes using hs8A element but the zebrafish/human atp11c/ATP11C promoters instead of the zebrafish gata2 promoter. Note similarity of pattern, but much weaker activation. Abbreviations: h: hindbrain; Tg: tegmentum; E: epiphysis; FP: floor plate; R: rhombomeres; Dc: diencephalon; tc: telencephalon; ie: inner ear; sc: spinal cord; C: cerebellum. |
|
The genomic locus of PAX6. (A) Ancora view of a 5 Mb window across the human locus (upper track), density of HCNEs compared with the mouse genome (lower track) and conserved synteny in the zebrafish genome. (B) 1 Mb mVISTA plot of human PAX6 compared with zebrafish Xenopus tropicalis, (xt, middle track) and Fugu rubripes (fr, lower track). HCNEs conserved in all four species were tested in zebrafish and are boxed in red. Insets above tracks show two enhancer detection insertions in the pax6a and pax6b genomic regulatory block in zebrafish and the RNA in situ hybridization pattern of these genes in a 2 day zebrafish embryo. Expression is mainly confined to the nervous system (for detailed expression see also Supplementary data). |
|
Lateral views of HCNE-test transgenic zebrafish embryos at 2 days post fertilization. Transgenes are labeled by locus name (pax6a/b/PAX6), organism (zebrafish = dr/human = hs) and HCNE number. (A, B) Show transgenes made with zebrafish HCNE 1 expressing GFP in optic tectum, pectoral fins and hindbrain vs. human element driving expression in dorsal root ganglia and hindbrain, which indicate its functional connection to WT1 expressed in these domains. HCNE 2 in (C, D) directs expression mainly to hindbrain with both zebrafish and human elements. Embryos in (E) made with human sequence PAX6_hs6 express in optic tectum, tegmentum, hindbrain, spinal chord and retina, embryo (F) expressing in dorsal diencephalon (PAX6_hs7). Sub-sequence pax6a_dr6A (as seen in Fig. 6B) drives expression only in optic tectum and retina but the human homologous PAX_hs6A element directs expression to optic tectum, hindbrain, spinal chord, lateral line and lens. |
|
Expression driven by PAX6 HCNE8, human and both zebrafish orthologous sequences. (A–C′) All sequences drive expression to pineal and diencephalon. Left column shows lateral views of 3 days old embryos, right column dorsal views of heads of the same larvae. The signals differ in relative intensity in these structures between human PAX6/pax6a/pax6b. (D) VISTA conservation plot of the alignment of zebrafish pax6a_dr8 and pax6b_dr8 with PAX6_hs8 (baseline) indicating asymmetrical divergence of the zebrafish duplicates. Common peak indicates core HCNE. |
|
Confocal study of PAX6 HCNE8. (A–F′) 3D reconstructions of dorsally-imaged confocal stacks of 3 dpf embryos, transgenic genotype indicated top right, stained using immunohistochemistry for GFP (green) combined with TOPRO nuclear staining (grey). Viewed from dorsal (A, B, C, D, E, F) and lateral (A′, B′, C′, D′, E′, F′) aspects. Areas of GFP expression in embryos bearing GFP transgenes driven by zebrafish pax6a (A, A′, B, B′) and pax6b (C, C′, D, D′) HCNE 8 were mapped onto schematic diagrams of the brain of a 3 dpf embryo drawn from dorsal (G) and lateral (with eye removed; G′) viewpoints as coloured areas: pineal = green, habenulae = blue, pretectal area = red, dorsal (roofplate) tectum = purple, dorsal tegmentum = yellow. 3D reconstructions were used to plot the locations of GFP-positive cells for multiple PAX6_hs8 embryos (n = 6) (examples E, E′, F, F′) using coloured circles. Where GFP-positive cells fell within the zones of either zebrafish HCNE 8-driven transgene expression the circle was coloured the equivalent colour. Where they fell outside these areas they were left white. The lateral schematic (G′) represents summated expression from both left and right sides of the brain, the black line across the image represents the depth of the 3D reconstructions employed (op = olfactory pits). |
|
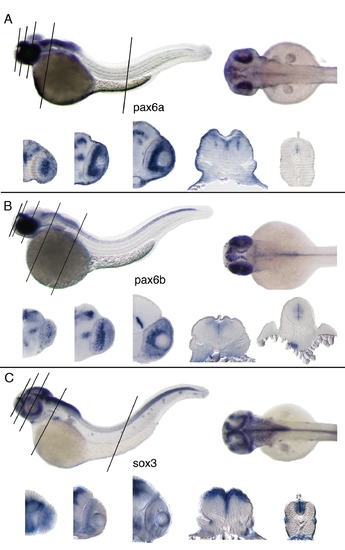
Lateral, dorsal views and vibratome sections of whole mount RNA in situ hybridizations. 2 days old zebrafish embryos and 20 μm thick transversal vibratome sections below (levels indicated by black lines on the whole mount). A: pax6a; expressed in hindbrain, optic tectum, anterior spinal cord, retina, diencephalon. B: pax6b; detected in hindbrain, spinal cord, retina, diencephalon, pancreas. C: sox3; detected in CNS, particularly in ventricular zone of the neural tube also in retina, branchial arches, lateral line an inner ear. |
|
Detection of bystander gene expression by RNA whole mount in situ hybridization. 1–3 days old zebrafish embryos. A–E: immp1l, a pax6b downstream bystander gene. Immp1l shows ubiquitous expression. B: rcn1, pax6b upstream bystander gene. Expression was detected in cartilage of the fins, also notochord, and weak expression in retina and facial skeleton; C: atp11c, the sox3 downstream bystander gene has low level expression, detected in the olfactory pits and lateral line. |
|
Zebrafish embryonic expression driven by SOX3_hs8 enhancer activating different minimal promoters: A: chicken β-globin promoter, B: zebrafish ngn 1, C: zebrafish hsp70, D: human SOX3, E: human ATP11C, F: zebrafish atp11c G: gata2 promoter, H: ATP11C-adjacent CTCF binding site and gata2 promoter. All patterns are confined to hindbrain, spinal cord, inner ear, olfactory pits and telencephalon (identical with pattern obtained with gata2 promoter used normally in our enhancer test assay). All transgenic embryos are 2 days old F1, lateral view. |
Reprinted from Developmental Biology, 327(2), Navratilova, P., Fredman, D., Hawkins, T.A., Turner, K., Lenhard, B., and Becker, T.S., Systematic human/zebrafish comparative identification of cis-regulatory activity around vertebrate developmental transcription factor genes, 526-540, Copyright (2009) with permission from Elsevier. Full text @ Dev. Biol.