- Title
-
Anti-apoptotic effects of IGF-I on mortality and dysmorphogenesis in tbx5-deficient zebrafish embryos
- Authors
- Tsai, T.C., Shih, C.C., Chien, H.P., Yang, A.H., Lu, J.K., Lu, J.H.
- Source
- Full text @ BMC Dev. Biol.
|
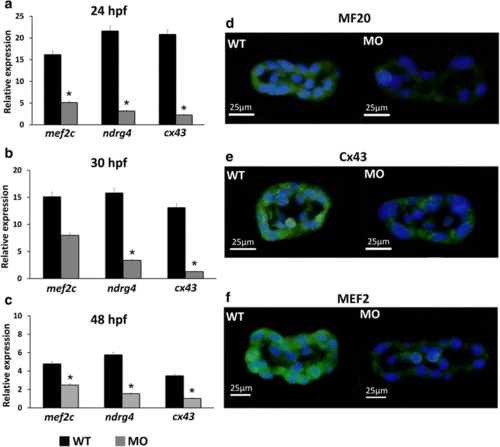
Expression levels of myocardium-related genes and cardiomyocyte proteins in wild-type embryos and tbx5 morphants. The MO group had significantly decreased expression of myocardium-related genes (mef2c, ndrg4, cx43) during the embryonic development stages (24 hpf (a), 30 hpf (b), and 48 hpf (c)) compared with the WT group. Zebrafish embryos at 30 hpf were stained with the myosin heavy chain antibody MF20 (green), connexin-43 antibody Cx43 (green), myocyte enhancer factor-2 antibody MEF2 (green) and counterstained with DAPI (blue) for nucleus identification. The MO group had decreased expression of MF20 (d), Cx43 (e), and MEF2 (f) compared to the WT group. Data are presented as the means ± standard deviations. *P < 0.05 vs. WT. WT, wild-type embryos; MO, tbx5 morphants. Hpf: hours post-fertilization. The number of specimens was 50, and the number of independent experiments was 3 in each group |
|
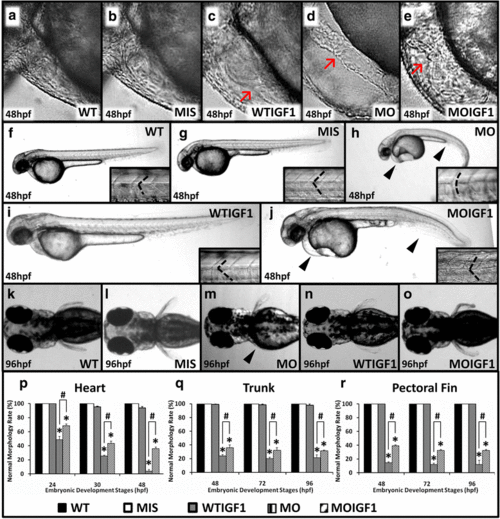
Phenotypes of the tbx5 morphants and IGF-I-treated tbx5 morphants. The WT (a) and MIS groups (b) had normal hearts, and the WTIGF1 (c) group had minimal pericardial effusion. The MO group (d) had string-like hearts accompanied by a massive pericardial edema, but the appearance of hearts in the MOIGF1 group (e) was improved. There were no significant differences observed in the trunks of the WT (f), MIS (g) and WTIGF1 groups (i), in which the trunks were straight and the somites appeared to be “V-shaped”. The trunk of the MO group (h) bent severely and showed a “U shape”. The MOIGF1 group (j) had partially-restored appearances of the trunks and curvatures of somites. The pectoral fins of the MO group (m) were truncated or undeveloped, but the WT (k), MIS (l), WTIGF1 (n), and the MOIGF1 group (o) had normal pectoral fins. The MO group had significantly lower normal morphology rates of the hearts (p), trunks (q) and pectoral fins (r). Compared with the MO group, the MOIGF1 group had significantly higher percentages of normal morphology rates of the hearts (p), trunks (q) and pectoral fins. No defective embryos were found in the WT and MIS group, and almost all of the WTIGF1 group embryos developed well. Red arrow: pericardial effusion; black arrow head: defect site; dashed line: shape of somite border. Data are presented as the means ± standard deviations. *P < 0.05 vs. WT; #P < 0.05 MOIGF1 vs. MO. WT: wild-type embryos; MO: tbx5 morphants; MIS: mismatch tbx5 morpholino-treated wild-type embryos; WTIGF1: IGF-I-treated wild-type embryos; MOIGF1: IGF-I-treated tbx5 morphants; hpf: hours post-fertilization. The number of specimens was 50, and the number of independent experiments was 3 in each group |
|
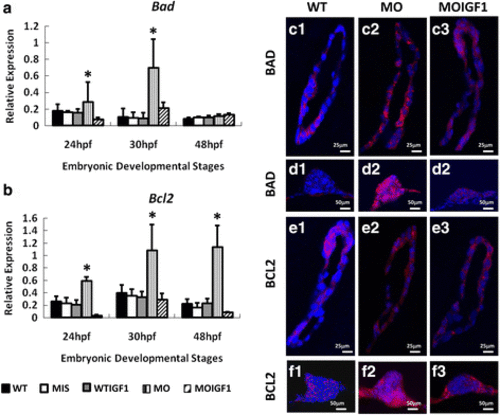
Expression of apoptosis proteins and genes in the tbx5 morphants and IGF-I-treated tbx5 morphants. Expression of the bad gene was significantly induced in the MO group (a) at 24 and 30 hpf, and the expression of bcl2 was significantly induced in the MO group at 24, 30 and 48 hpf. b. There were no significant differences in the expression levels of bad and bcl2 between the WT, WTIGF1 and MOIGF1 groups. Zebrafish embryos at 30 hpf were stained by the proapoptotic-related antibody BAD (red) and anti-apoptotic related antibody BCL2 (red) and were counterstained with DAPI (blue) for nuclear observation. Expression levels in the sagittal sections of hearts are shown in C1–3 and E1–3; expression levels in the transverse sections of pectoral fins are shown in D1–3 and F1–3. The WT group had very low expression of the BAD apoptotic protein (Figure 6C1, 6D1) and BCL2 pro-apoptotic protein (Figure 6E1, 6F1). Expression of the BAD apoptotic protein (Figure 6C2, 6D2) and BCL2 pro-apoptotic protein (Figure 6E2, 6F2) was induced significantly in hearts and pectoral fins in the MO group, whereas expression of BAD (Figure 6C3, 6D3) and BCL2 (Figure 6E3, 6F3) was significantly reduced in the MOIGF1 group. Data are presented as the means ± standard deviations. *P < 0.05 vs WT. WT: wild-type embryos; MO: tbx5 morphants; MIS: mismatch tbx5 morpholino-treated wild-type embryos; WTIGF1: IGF-I-treated wild-type embryos; MOIGF1: IGF-I-treated tbx5 morphants; hpf: hours post-fertilization. The number of specimens was 50, and the number of independent experiments was 3 in each group |
|
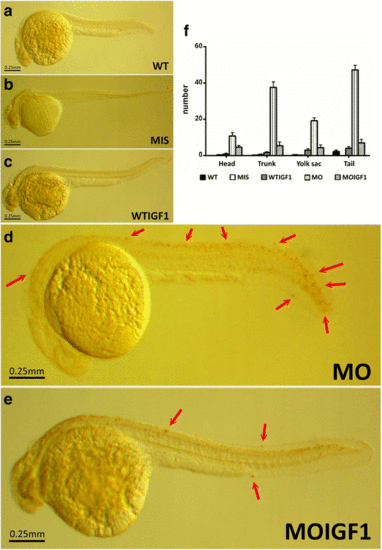
TUNEL assay for the tbx5 morphants and IGF-I-treated tbx5 morphants. TUNEL assay revealed no TUNEL-positive cells in the WT (a), MIS (b), and WTIGF1 (c) groups. However, massive numbers of TUNEL-positive cells were visible in the MO group (d). In the MOIGF1 group, the amount of TUNEL-positive cells was reduced (e). Most of the TUNEL-positive cells were clustered over the tail and trunk, while several were be found over the yolk sac and head (f). Arrow, TUNEL-positive cells. Data are presented as the means ± standard deviations. WT: wild-type embryos; MO: tbx5 morphants; MIS: mismatch tbx5 morpholino-treated wild-type embryos; WTIGF1: IGF-I-treated wild-type embryos; MOIGF1: IGF-I-treated tbx5 morphants. The number of specimens was 10, and the number of independent experiments was 3 in each group |