- Title
-
Genome editing with RNA-guided Cas9 nuclease in Zebrafish embryos
- Authors
- Chang, N., Sun, C., Gao, L., Zhu, D., Xu, X., Zhu, X., Xiong, J.W., and Xi, J.J.
- Source
- Full text @ Cell Res.
|
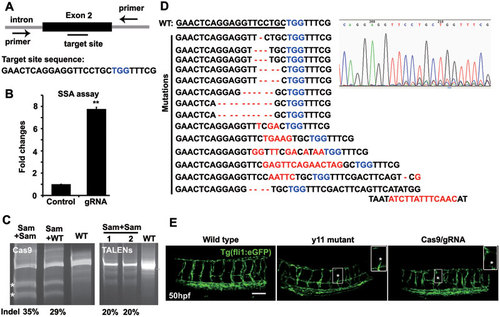
Cas9/gRNA induces indels in the etsrp locus in zebrafish. (A) Cartoon showing the position of the target site and its sequence in the etsrp locus in zebrafish. (B) The SSA recombination assay showing luciferase activity with and without cleavage of the target site by the Cas9/gRNA system in 293T cells. The luciferase activity increased by 8-fold in the group with gRNA (Cas9 + gRNA) compared with the control group (Cas9 + empty vector). Error bars indicate SD, n = 3. **P < 0.01. (C) Representative SURVEYOR assay showing 35% efficiency of Cas9-mediated cleavage in a single embryo and 20% by TALENs-mediated cleavage in 2 embryos at 50 hpf. The indels of 8 embryos were measured with a frequency of 31.2% - 38.4%. * indicates the cleavage bands. Sam, sample treated by Cas9/gRNA or TALENs; WT, wild type. (D) Representative Sanger sequencing results of the PCR amplicons from 8 individual embryos at 50 hpf showing indels (red) induced by Cas9/gRNA in the targeted etsrp locus. PCR fragments were amplified and cloned into the pEASY vector from an individual embryo for Sanger sequencing (20-30 clones for each embryo). TGG (blue) is the PAM sequence. Deletion is represented by a dashed line and insertion is highlighted in red. (E) Fluorescent images showing abnormal intersegmental vessels that were similar in the etsrpy11 mutant and Cas9/gRNA-induced mutant, compared with the wild-type transgenic embryo at 50 hpf. * indicates defects in intersegmental vessel sprouting that are enlarged in the insets. Scale bar, 100 μm. |
|
Cas9/gRNA induces indels in the gata5 and gata4 loci in zebrafish. (A) Cartoon showing the position of the gRNA-targeting site and its sequence in the gata5 locus in zebrafish. (B) SSA recombination assay showing luciferase activity with and without cleavage of the target site by the Cas9/gRNA system in 293T cells. The luciferase activity increased by 16-fold in the group with gRNA (Cas9 + gRNA) compared with the control group (Cas9 + empty vector). Error bars indicate SD, n = 3. **P < 0.01. (C) Representative SURVEYOR assay showing the efficiency of Cas9-mediated cleavage, 26.4% in a single embryo at 50 hpf. (D) Representative Sanger sequencing results of the PCR amplicons of 8 individual embryos at 50 hpf, showing indels induced by Cas9/gRNA in the targeted gata5 locus. Twenty to thirty clones were sequenced for each embryo. Table summarizes the frequencies of site-specific indels, ranging from 50% (11/20) to 90% (18/20), in 8 individual embryos tested by Sanger sequencing. TGG (blue) is the PAM sequence. Deletion is represented by a dashed line and insertion is highlighted in red. (E) Two small hearts were formed in a Cas9/gRNA-induced mutant (right), which phenocopied that in the fautm236a genetic mutant, whereas a single heart formed in control embryos at 60 hpf. The heart was stained with phalloidin. * indicates two small hearts. Scale bar, 100 μm. (F) Representative Sanger sequencing results of the PCR amplicons of 8 individual embryos at 50 hpf, showing indels induced by Cas9/gRNA in the targeted gata4 locus as in D. AGG (blue) is the PAM sequence. AGG in 2 out of 16 sequencing results was deleted. PHENOTYPE:
|

Unillustrated author statements PHENOTYPE:
|